3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2014; 11(7):732-741. doi:10.7150/ijms.7768 This issue Cite
Review
Paratyphoid Fever: Splicing the Global Analyses
1. Department of Medical Microbiology, Faculty of Medicine, University of Malaya, 50603 Kuala Lumpur.
2. Department of Biomedical Science, Faculty of Medicine, University of Malaya, 50603 Kuala Lumpur.
3. Institute of Biological Sciences, Faculty of Science, University of Malaya, 50603 Kuala Lumpur.
Received 2013-9-27; Accepted 2014-3-5; Published 2014-5-14
Abstract
The incidence of enteric fever caused by Salmonella enterica serovar Paratyphi A (S. Paratyphi A) is increasing in many parts of the world. Although there is no major outbreak of paratyphoid fever in recent years, S. Paratyphi A infection still remains a public health problem in many tropical countries. Therefore, surveillance studies play an important role in monitoring infections and the emergence of multidrug resistance, especially in endemic countries such as India, Nepal, Pakistan and China. In China, enteric fever was caused predominantly by S. Paratyphi A rather than by Salmonella enterica serovar Typhi (S. Typhi). Sometimes, S. Paratyphi A infection can evolve into a carrier state which increases the risk of transmission for travellers. Hence, paratyphoid fever is usually classified as a “travel-associated” disease. To date, diagnosis of paratyphoid fever based on the clinical presentation is not satisfactory as it resembles other febrile illnesses, and could not be distinguished from S. Typhi infection. With the availability of Whole Genome Sequencing technology, the genomes of S. Paratyphi A could be studied in-depth and more specific targets for detection will be revealed. Hence, detection of S. Paratyphi A with Polymerase Chain Reaction (PCR) method appears to be a more reliable approach compared to the Widal test. On the other hand, due to increasing incidence of S. Paratyphi A infections worldwide, the need to produce a paratyphoid vaccine is essential and urgent. Hence various vaccine projects that involve clinical trials have been carried out. Overall, this review provides the insights of S. Paratyphi A, including the bacteriology, epidemiology, management and antibiotic susceptibility, diagnoses and vaccine development.
Keywords: Salmonella Paratyphi A, paratyphoid fever, epidemiology, antibiotic resistance, diagnosis, vaccine.
Introduction
Enteric fever is still an important public health problem in many developing countries. It is difficult to estimate the real impact of this disease as the clinical symptoms may be confused with other febrile illnesses and specific laboratory confirmation may not be available in these areas (1). Enteric fever is a multi-system disease characterized by prolonged fever, sustained blood stream infection, activation of the endothelial system, metastatic infections and immunologic complications due to immune complex deposition leading to multi-organ dysfunction (2). Overall, enteric fever is caused by Salmonella enterica serovar Typhi (S. Typhi) and Salmonella enterica serovar Paratyphi A (S. Paratyphi A), B and C. However, S. Paratyphi A has begun to replace S. Typhi as the main causative agent of enteric fever in many Asian countries in recent years. The disease caused by S. Paratyphi A is well known as paratyphoid fever. The incidence of paratyphoid fever is increasing gradually worldwide (3), especially in certain endemic regions, such as certain provinces in China and Pakistan where S. Paratyphi A infection has become a major health problem (4). In 2000, 5.4 million cases of paratyphoid fever caused by S. Paratyphi A were estimated (5). The disproportionate increase in the numbers of S. Paratyphi A cases may be due to the vaccine effect (Ty21a and Vi vaccines), which only protects individuals from S. Typhi infection (5, 6) and also the inappropriate preventive strategies against S. Paratyphi A. The reduction strategies that were effective against S. Typhi might not be useful against S. Paratyphi A (3).
S. Paratyphi A can be isolated from the blood and faeces from paratyphoid fever patients (7). This bacterium causes a milder infection with lower mortality and chronic carriage rate compared to S. Typhi (8). Transmission of the pathogen is via the fecal-oral route, with humans as the sole reservoir of infection. Transmission is via consumption of contaminated food/water or contact with chronic asymptomatic carriers (4). Family contact could be a factor of transmission as well, but a recent report on the risk factors for the disease showed that paratyphoid infections occur mostly outside the household (9). The important vehicles of transmission in many countries include shellfish harvested from sewage-contaminated beds, raw fruits, vegetables fertilized by night soil and eaten raw, milk and milk products. Preparation of food by hands of carriers or infected food handlers may also contribute to the disease transmission (10).
Symptoms of S. Paratyphi A infection include fever, headache, diarrhoea or constipation, malaise, anorexia, nausea, dry cough, gastrointestinal symptoms, abdominal pain, chills, raised spots or rashes on body (9, 11). Overall, the cytokine profile of S. Paratyphi A infection is similar to S. Typhi but distinct from other non-typhoidal Salmonellae. During the acute phase of infection, IFN-γ is remarkably induced in addition to the increase of IL-6, IL-8, IL-10, IL-15, and TNF-α. However, the white blood cell count of paratyphoid fever patients does not increase significantly during the acute phase as opposed to other infectious diseases (12).
Paratyphoid fever may evolve into a carrier state. A carrier state is defined as the shedding of Salmonella in the stools or urine after resolution of acute illness, while the chronic carrier state is defined as the long-term excretion of Salmonella in the stools for more than one year and this occurs in less than 5% of the patients with enteric fever (8, 13, 14). Chronic carriers are at particular risk of transmitting infection to others, especially if they are food-handlers (14). The identification for persistent excreters of S. Typhi and S. Paratyphi A is important to prevent transmission of the pathogen to others. However, the excretion of pathogenic organisms by chronic carriers may be intermittent. So, no practical system of microbiological clearance or screening could identify all chronic carriers (8).
Chronic biliary carriage may occur in 2 - 5% of cases, even after treatment. Biliary carriage is defined as continued shedding of the organism for more than a year, and is a public-health risk, especially for infected individuals who work in the food industry (6).
In this paper, we aimed to provide a better understanding of S. Paratyphi A, the causative agent of paratyphoid fever from the aspects of microbiology, genome composition, global trend of epidemiology, management, diagnoses and vaccine development.
Bacteriology
Generally, S. Paratyphi A is a Gram-negative bacterium that belongs to the Enterobacteriaceae family (15). Based on the serotyping scheme developed by Kauffman-White, S. Paratyphi A is classified as serogroup A with an antigenic formula as 1,2,12:a:-. S. Paratyphi A (1,2,12:a:-) resembles S. Sendai 1,9,12:a:1,5) as both are poor Hydrogen Sulfide (H2S) producers (7). Howere, they can be distinguished by the ability to ferment xylose. According to Edwards and Ewing (16), S. Paratyphi A produces a weakly positive reaction for H2S test and subsequently fails to show evidence of H2S production during the first of 14 days of incubation. Gas produced by S. Paratyphi A is also relatively little and S. Paratyphi A is not lysine decarboxylase-positive organism. (7).
The Genome features of S. Paratyphi A
Two genomes of S. Paratyphi A, ATCC 9150 and AKU 12601 were sequenced and compared with S. Typhi previously by McClelland et al. (17), Didelot et al. (18) and Holt et al. (19). The genome size of ATCC 9150 is 4,585,229 bp encoding 4,263 CDS while AKU12601 is 4,581,797 bp in size and encodes for 4,285 CDS. Both S. Paratyphi A genomes are smaller than S. Typhi CT18 and Ty2 (17, 19).
Both genomes of S. Paratyphi A ATCC 9150 and AKU 12601 harbor three phages and 39 insertion/deletion events and 188 SNPs (dN/dS ratio of 0.62) have been identified (19). Unlike ATCC 9150, AKU 12601 harbours a multidrug resistant plasmid, IncHI1 which is approximately 212,711 bp. The presence of plasmids associated with multidrug resistance in S. Paratyphi A strains has been reported by Holt et al. (19), Mandal et al. (20) and Panigrahi et al. (21). Holt et al. (19) reported that the IncHI1 plasmids in S. Typhi and S. Paratyphi A are highly similar. This is probably due to transfer of plasmids between the two serovars (19). Besides that, isolation of cryptic plasmid in a clinical strain of S. Paratyphi A was also reported by Huang et al. (22).
More recently, Liang et al. (23) has sequenced five clinical and environmental strains of S. Paratyphi A isolated from Zejiang, Guizhou, Jiangxi, Guangxi and Yunnan province, respectively. These draft genomes were compared with the two reference strains, ATCC 9150 and AKU12601. In their comparative genomic analyses, 4252 orthologs have been identified. Among the anthologies, 3720 genes were derived from core genomes, while 465 genes were dispensable. Only 67 were strain-specific genes. This suggests that the genome of S. Paratyphi A is highly conserved (23). However, mutations may have been introduced by recombination that favours the adaptation of the strains in their ecological niche (18, 23). Most of the pseudogene clusters were homologous to functional gene clusters and increasing number of pseudogene clusters could lead to the inactivation of functional genes (23).
Epidemiology
S. Paratyphi A is increasingly important as the causative agent (50% of Salmonella bloodstream isolates) of enteric fever in Asia (5). It is the second leading cause of enteric fever in Asia, the Middle East, Africa and South America after S. Typhi (21) (Figure 1).
Distribution of S. Paratyphi A in different countries in Asia and Europe.
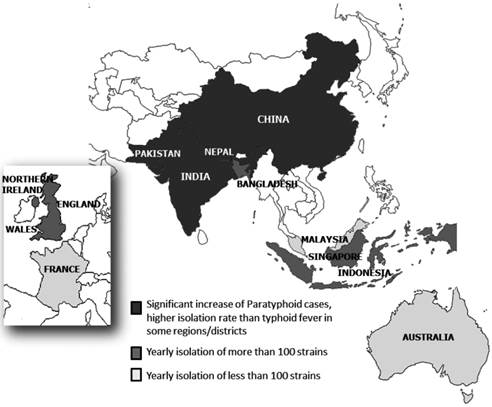
South Asia
In India, no major outbreak of paratyphoid fever has been recorded, although it was implicated to cause 3% - 17% of enteric fever cases before October 1995. However, Kumar et al. (24) revealed that among the enteric fever cases that occurred in an urban slum in New Delhi from October 1995 to October 1996, 25% was caused by S. Paratyphi A. Moreover, in 1996, 36 cases of paratyphoid fever were reported in a residential area of New Delhi, India within 1 month period (September - October) (25, 26). Subsequently, infections due to S. Paratyphi A are increasing in India. A few retrospective cohort studies have been conducted in India to monitor the trends of S. Paratyphi A infection. Based on the data collected by Sood et al. (27) from the All India Institute of Medical Sciences in New Delhi, the isolation rate of S. Paratyphi A has increased from 6.5% in 1994 to 44.9% in 1998. Data from 1999 to 2004 were collected again from the same institution by Mohanty et al. (28) and the isolation rate was dropped to 23.8%. More recently, in another study conducted by Capoor et al. (28), a significant increase in S. Paratyphi A isolation from 2001 to 2006 has been observed in New Delhi.
Unusual increase of S. Paratyphi A infection has also been observed from other regions of India. For example, S. Paratyphi A caused 59% of total enteric fever cases in Calicut (now known as Kozhikode) in 2003 (30), 23.5% in Calcutta from September 2003 to August 2004 (3), 40.6% in Chandigarh in 2007 (31), 20.3% in Mumbai in 2002 (32), 38.4% in Shimla from 2000 to 2006 (33), 23.3% in Chennai between 2007-2009, and the isolation rate of S. Paratyphi A in Nagpur and Sevagram was 46.2% (34) and 53.3% (35), respectively between 2001 to 2003. Moreover, the isolation of S. Paratyphi A was relatively higher than S. Typhi in Bangalore. In the year 2004, 79 S. Typhi and 170 S. Paratyphi A isolates were isolated from patients suffering from enteric fever at the Manipal Hospital (36). More recently, a multi-center surveillance study has been carried out by the Indian Network for Surveillance of Antimicrobial Resistance Group. A total of 764 S. Paratyphi A strains have been isolated between January 2008 to December 2010 in the 15 participating centers throughout India (37).
S. Paratyphi A infection had emerged in Pakistan and Nepal. In Pakistan, there was a significant increase of S. Paratyphi A infection in Rawalpindi (16, 38) and Karachi since 1996 (3, 39) while in Nepal, a higher isolation rate of S. Paratyphi A during summer has been documented (40, 41). However, a higher prevalence of S. Paratyphi A compared to S. Typhi, (with the ratio of 39:30 from a total of 69 isolates) was also observed in autumn (September and October) as reported by Shirakawa et al. (42). In Patan Hospital, Kathmandu, 2677 S. Paratyphi A strains were isolated from blood of patients (accounted for 29.3% of enteric fever) between 1993 and 2003 (43). Later, during January-August 2004, the isolation rate of S. Paratyphi A increased and accounted for 32.8% of enteric fever in the same hospital (43). Between June 2005 and May 2009, the isolation rate of S. Paratyphi A was slightly decreased and accounted for 31.5% out of 3,898 cases of blood culture-confirmed enteric fever (44). Similarly, 288 out of 541 blood culture samples from patients with enteric fever collected in Tribhuvan University Teaching Hospital, Kathmandu between January and September, 2004 were serotyped as S. Paratyphi A (45).
In Bangladesh, prevalence of paratyphoid fever is still not as high as typhoid fever. In Dhaka, Bangladesh, 8 paratyphoid fever cases were detected among 48 enteric cases reported for the year 2003 (46). In another study carried out by Sheikh et al. (47), only 5 out of 89 patients with enteric fever were confirmed as paratyphoid fever cases.
East Asia
In China, the incidence of paratyphoid fever has also increased rapidly as reported in San Jiang county and Quanzhou county in 1995 (48). The predominance of S. Paratyphi A infections has superceded S. Typhi in most parts of China. During 1994-2006, a total of 1855 paratyphoid fever cases were reported by the Center for Disease Prevention & Control (GXCDC), China. More paratyphoid cases were noticed as compared to typhoid cases from August 2001 to July 2002 in Heichi City while S. Paratyphi A was responsible for all the enteric fevers between April 2006 to December 2007 in Quanzhou city. In Shenzhen, 70.3% of 91 patients in Shenzhen People's Hospital were diagnosed with S. Paratyphi A infection (49) during the period of 2002 to 2007. Based on the study by Wu et al. (49), more paratyphoid fever cases were observed in Shenzhen People's Hospital in 2003. In Hong Kong, paratyphoid fever was less common than typhoid fever. However, the cases of paratyphoid fever have exceeded typhoid fever in the year 2003, where 60 cases of paratyphoid fever were recorded as compared to 49 typhoid fever cases (50). This indicates the wide dispersal of S. Paratyphi A in this region during 2003.
Southeast Asia
In Indonesia, S. Paratyphi A is increasingly important, but the infection rate is still lower than the S. Typhi. During August 2002- July 2003 surveillance, 154 enteric fever cases were reported in Jakarta, but only 14% were confirmed as S. Paratyphi A infection (3). The same findings were also noted in another study by Vollaard et al. (9) which showed more S. Typhi than S. Paratyphi A within the period of June 2001 to February 2003. These studies indicated that S. Typhi remains the main cause of enteric fever in Jakarta, Indonesia.
In Singapore, outbreaks of paratyphoid fever were mainly due to imported food. In 1979, there were 61 laboratory-confirmed S. Paratyphi A cases in an outbreak and imported fresh oysters were confirmed as the vehicles of transmission (51). The largest outbreak happened in 1996 where 167 cases of S. Paratyphi A infections were reported between February and May where imported de-shelled coconut was suspected as the vehicle of transmission (52). During the 19 years period (1990-2009), 2464 enteric fever cases were notified and among these cases, 707 were caused by S. Paratyphi A (259 indigenous cases and 448 imported cases) (53).
In Malaysia, the national Salmonella surveillance data are collected through passive surveillance of laboratory-confirmed human Salmonella isolates. Based on the study by Jegathesan (54) and Md. Yasin (55), S. Paratyphi A was uncommon compared to S. Typhi. However, the number of isolates has increased from 169 (1973-1982) to 180 (1983-1992).
Australia
Since 2003, there was an upsurge in S. Paratyphi A infection in Australia. However, the confirmed paratyphoid fever cases were often associated with travel. Among 810 S. Paratyphi A isolated between 1985-2010, 547 isolates were originated from India, Indonesia, Bangladesh, Pakistan, Nepal, Cambodia, Thailand, Philippines, Papua New Guinea and Lebanon (56). In another study carried out in Sydney, 8 S. Paratyphi A infections were detected during the period January-June 2011 and the patients were predominantly associated with travels to the Indian subcontinent (57).
Europe
It is undeniable that there is a risk of disease transmission across different geographic regions and paratyphoid fever is now known as 'travel-associated' disease (6). Threlfall et al. (58) also cautioned the choice of first-line drugs for treatment of infections with S. Typhi and S. Paratyphi A in European countries as treatment failure might occur in patients especially for those who have a record of travels to areas where drug resistant strains are endemic.
S. Paratyphi A remains the main cause of paratyphoid fever in England, Wales and Northern Ireland and there was an average of 192 paratyphoid cases reported each year from 1995 to 2005. In 2005, 61 cases of S. Paratyphi A infection were associated with travels to India, and 31 travels to Pakistan (59). Later, between 1 May 2006 and 30 April 2007, 224 S. Paratyphi A infections have been reported and 100% of the patients had travel histories (60). Overall, the rate of S. Paratyphi A isolates was slightly higher than S. Typhi before 2007, however, S. Typhi has become predominant since 2008 (ratio of S. Paratyphi A to S. Typhi from 2008 to 2012 were 237:268, 185:248, 212:287, 223:261 and 167:177, respectively) in England, Wales and Northern Ireland (60, 61, 62, 63, 64). In France, there were only 16 cases of paratyphoid fever caused by S. Paratyphi A from 1988 - 1998. Among the 16 patients, 7 of them had been vaccinated against typhoid fever and this proves that current typhoid vaccines might not provide full protection against S. Paratyphi A infection (65).
Management of S. Paratyphi A infection and the increase of antibiotic resistance
Studies on age-related clinical and microbiological characteristic of enteric fever have been carried out. Walia et al. (66) and Teoh et al. (52) reported age-related predilection of paratyphoid fever among adults while Khuribulos (67) reported the high prevalence of paratyphoid fever among children younger than 5 years of age. According to Vollaard et al. (9), the age of patients who suffered from paratyphoid fever did not differ significantly from typhoid fever patients. However, transmission of the pathogen from adults to children could occur for those who are ignorant of personal hygiene. Therefore, awareness of personal and environmental hygiene is very important, especially in those endemic countries. A study to evaluate the effectiveness and efficiency of public health management of cases of infection due to S. Typhi and S. Paratyphi A from 2002 to 2004 had been carried out in Northeast London. No chronic carrier of S. Paratyphi A was found. However, the current guidelines and practice was not sufficient for case follow-up and contact screening (8).
Today, most paratyphoid fever occurs in less developed countries where sanitary conditions remain poor and the water supplies are not treated (6). Obtaining accurate data on the occurrence of disease in these countries is also difficult because the diagnosis of paratyphoid fever is often based on clinical assessment, without blood culture confirmation and most patients are treated as outpatients (11). In Jakarta, Indonesia, over 80% of patients with typhoid or paratyphoid fevers are treated as outpatients (9). Most of the patients are not aware that they might become infected during the first week of convalescence without any symptoms. Without any treatment, the bacteria will remain in one's body and will be discharged for up to three months. Disease transmission can occur during this period if such infected individuals are involved in food handling or lack of personal hygiene (4). Sometimes, a patient could be infected by both S. Typhi and S. Paratyphi A (68, 69).
Infections with S. Paratyphi A and S. Typhi are commonly treated with ciprofloxacin (70). However, increasing multidrug resistant strains of S. Paratyphi A and decreasing ciprofloxacin susceptibility have been reported since the 1990s (70). S. Typhi and S. Paratyphi A with decreased susceptibility to fluoroquinolones and resistance to nalidixic acid are commonly reported in India, Pakistan, Japan, China and Southeast Asia (49, 70, 71, 72, 73). However, nalidixic acid and ciprofloxacin resistance was more commonly seen in S. Paratyphi compared to S. Typhi (68). Shirakawa et al. (42) had reported a high mutation rate of gyrA gene in S. Paratyphi A and such strains are resistant to nalidixic acid. Mutations in the gyrA gene that lead to quinolone resistance and reduced susceptibility to fluoroquinolones are clinically significant in S. Paratyphi A. Although azithromycin has been found to be efficacious for the treatment of uncomplicated typhoid fever (74, 75, 76, 77, 78, 79), high azithromycin MIC value and a case of azithromycin treatment failure in a patient with invasive S. Paratyphi A infection have been reported (80, 81).
Clinical presentation and diagnosis of paratyphoid fever
Clinical diagnosis of paratyphoid fever can be difficult because the symptoms are not unique and overlap with other febrile illness, especially malaria and dengue. In addition, both typhoid and paratyphoid fever share the same symptoms and it is difficult to differentiate these two diseases (9).
Basically, paratyphoid fever has three clinical stages: an early stage marked by high fever; a toxic stage with abdominal pain and intestinal symptoms, and a long period of recovery stage of fever or defervescence. Toxic stage is the most important stage as there is a 1-10% chance of intestinal perforation, hemorrhage or inflammatory destruction (9). The infection may develop cardiac complications and sometimes fatal in adults and children (82, 83, 84, 85). Early identification of the specific etiological agent and knowledge of local antimicrobial resistance patterns would be invaluable in guiding rational treatment decisions (86).
In many endemic countries, symptoms or combine-symptoms are the main tools for diagnosis of paratyphoid infection. However, a few case studies had shown that these symptoms should not be the standard for paratyphoid fever's diagnosis because most of these symptoms are too common (87, 88, 89). In general, patients with paratyphoid fever have more rose spots than patients with typhoid fever. However, rose spots are absent sometimes or not apparent in dark-skinned patients (9, 88). For example, a man of Indian origin was admitted to a hospital in UK because of fever and severe headache with chills. Differential diagnoses were carried out on that patient, such as diagnosis for malaria, dengue fever, and meningitis. Diagnosis of typhoid or paratyphoid fever was not considered because of the absence of rose spot. However, growth of S. Paratyphi A in blood culture confirmed that the patient was indeed infected with S. Paratyphi A (88).
In general, the mortality and morbidity rates for paratyphoid fever are much lower than typhoid fever (8, 14). Liver abscesses due to S. Paratyphi are extremely rare. However, liver abscesses caused by S. Paratyphi A infection were reported by Jeans and Mckendrick (89). A man who had suffered from 8 days of serious abdominal pain, fevers, diarrhoea, and nausea was admitted to the hospital. Abdominal computerised tomography scan result showed a 6 cm mass within the right lobe of the liver. Confirmatory diagnosis was carried out with serological test and the test strongly suggested that the patient had an amoebic liver abscess with secondary infection by S. Paratyphi A (89).
Current laboratory detection of S. Paratyphi A
Paratyphoid fever could not be distinguished clinically from typhoid fever. Consequently, due to the limited sensitivity of symptom-combinations, the clinical presentation cannot be used as a screening method. Laboratory tests are essential for the confirmation of paratyphoid fever (9).
Generally, culture method and serological test are the two main conventional laboratory diagnoses for S. Paratyphi A. However, in many countries, both laboratory methods are combined to identify an infection (38). Firstly, blood samples are collected from patients clinically suspected of having typhoid or paratyphoid fever. Blood culture is then performed. Finally, serological test using patient's serum is performed to confirm the infection of S. Typhi or S. Paratyphi A. The whole process requires more than one week for the final identification (38). However, when both culture and serology methods are applied, it increases the chance of detecting S. Paratyphi A specifically.
Culture method
Culture method is defined as the isolation of a bacterium from clinical specimens such as blood, bone marrow, stools, urine and intestinal secretions (14). Culture method for the detection of S. Paratyphi A is similar to other Salmonellae and S. Typhi based on the isolation on selective media such as Salmonella-Shigella agar (SS), xylose-lysine-desoxycholate agar (XLD) followed by identification using standard biochemical reactions (90). Detection of S. Paratyphi A by culture method is time consuming and usually requires 5-11 days (10).
There are many limitations in culture-based methods. The accuracy of diagnosis depends on the adequate amount of sample taken from patients, the appropriate media to be used, the stage of disease, and other variables during the isolation procedure (11). In typhoid fever, the number of bacteria is reported to be low in patients' blood and declined within the duration of illness (90), hence the sensitivity of diagnostic tools is varied during different stages of infection. Unlike typhoid fever, the number of bacteria throughout the duration of paratyphoid fever has not been reported previously. Therefore, this makes the diagnosis of paratyphoid fever more challenging. However, for both typhoid and paratyphoid fever, culture from the bone marrow gives the most accurate result (76).
Serological Method
Serology method is performed for diagnostic purposes when an infection is suspected. The widely used serological diagnostic modality for typhoid and paratyphoid fever is the Widal test which detects antibodies against the O-somatic and H-flagellar antigens (a 4-fold rise between acute and convalescent sera) in clinical specimens of suspected patients. It is commonly used throughout the world for the diagnosis of S. Paratyphi A infection (91).
However, Widal test is not very accurate and insensitive because of cross-reactivity with other bacteria which can also react with the antigens (91, 92). Furthermore, individuals who have prior typhoid vaccination may also give a false positive result (6). Nevertheless, Widal test remains the most widely used method for serological diagnosis of paratyphoid fever especially in the developing countries because it is inexpensive and easy to perform (92, 93).
More recently, Tam et al. (94) had developed a colorimetric test (TUBEX-PA) for paratyphoid fever. This test targeted lipopolysaccharides of S. Paratyphi A and could produce the result in 5 minutes. However, this detection system was less sensitive than culture-based detection as it could also detect more than 50% of typhoid patient in the study conducted (94).
Molecular Method for S. Paratyphi A detection
McClelland et al. (17) had reported that S. Typhi and S. Paratyphi A are genetically identical, and these similarities may be exploited to differentiate both infections. By sequencing the genome of S. Paratyphi A and comparing it to the genome of S. Typhi, researchers found that the pathogens have evolved along similar path. Comparative genomic hybridization (CGH) experiments and phylogenetic analysis of Salmonella serovars have also demonstrated the genetic relatedness of serovars Typhi and Paratyphi A, even though they are members of different serogroups (serogroup D1 and A, respectively) (95).
The advances in molecular biology have contributed greatly to the diagnosis of infectious diseases. Techniques which are based on the detection of the nucleic acids of pathogens are highly specific and sensitive compared to phenotypic methods. There are two major molecular approaches used in the detection of pathogens, such as nucleic acid hybridization and Polymerase Chain Reaction (PCR) (96).
PCR technique has provided increased sensitivity, allowed for more rapid processing times and enhanced the likelihood of detecting bacterial pathogens because it amplifies target DNA sequences that are present (86).
Most of the studies today manage to identify the bacteria up to serovar level. PCR technique has also been widely applied for the detection of Salmonella spp. However, the specific detection for S. Paratyphi A is relatively rare compared to S. Typhi. Hirose et al. (97) had successfully developed a multiplex PCR, which targeted different genes of S. Typhi and S. Paratyphi A such as rfbE, rfbS, viaB, and fliC. Tracz et al. (95) also developed a genomic approach which is able to detect S. Typhi and S. Paratyphi A.
Ou et al. (98) had successfully developed an alternative multiplex PCR assay to detect S. Paratyphi A. This assay was based on bioinformatics-led translational genomic approach and consists of 4 pairs of primers which work together in identifying S. Paratyphi A and most importantly to distinguish S. Paratyphi A from S. Typhi and other Salmonella serovars. Further evaluation of this assay was carried out by Teh et al. (99) and reported that this assay is specific for S. Paratyphi A identification.
Recently, Nga et al. (100) had developed a multiplex Real-time PCR assay to detect S. Typhi and S. Paratyphi A from biological specimens. This multiplex real-time PCR was evaluated in blood and bone marrow samples. Overall, specificity of this assay on various biological specimens is high (100%), however limited sensitivity on blood samples was observed.
Vaccines and future prospect
In early 1960s, a combined formulated vaccine, TAB was used to protect against typhoid and paratyphoid fever. The efficiency of this vaccine is 90%, and the protection period is as long as 5 years. However, this vaccine has not been widely used (101). In the past decades, only Ty21 and Vi polysaccharide vaccine that give protection against S. Typhi are available (1, 76).
Typhoid vaccination programs in Thailand, China, Vietnam, and India had allowed the emergence and increase of paratyphoid fever (4, 6, 76). For example, in Thailand, this vaccination program had led to a decrease of S. Typhi but had no effect on S. Paratyphi A. Similarly in other parts of the world, the disproportionate increase in the numbers of cases of S. Paratyphi A may be due to a vaccine effect, which gives protection only for S. Typhi (Ty21a and Vi vaccines) (4,6).
The development of vaccine for S. Paratyphi A remains a continuous effort. National Institutes of Health, Bethesda, MD had developed a new vaccine composed of surface-O-specific polysaccharide conjugated to tetanus toxoid as described by Konadu et al. (102). This vaccine is able to elicit IgG antibodies with bactericidal activity in the serum of patients infected with S. Paratyphi A. Field trials were carried out in Vietnam targeting all different age groups such as adults, teenagers, and 2- to 4-year-old children. As the outcome of phase I and II clinical trial were satisfactory and shown to be safe, a phase III clinical trial is planned in China (102).
On the other hand, Roland et al. (103) and Gat et al. (104) have been developing live attenuated S. Paratyphi A vaccines as the live attenuated vaccines provide dosing convenience and the induction of strong humoral, mucosal and cellular immunity (103). In the study of Gat et al. (104), the role of flagellar protein for immunity and protection has been explored while Roland et al. (103) had created phoPQ mutant as strong candidates for S. Paratyphi A vaccine. More recently, Micoli et al. (105) has described a new conjugation chemistry to developed O:2-CRM197-based conjugate vaccines. The conjugates, O:2-ADH-SIDEA-CRM197 and O:2-CDH-SIDEA-CRM197 are immunogenic in mice and generates good antibodies responses against S. Paratyphi A.
Conclusion
The high prevalence of S. Paratyphi A and the emergence of multidrug resistant clone are of great concern. Availability of reliable diagnostic tools and vaccines undoubtedly will ease the global burden, but might not be feasible in countries where a high proportion of people are living in poverty. As the ultimate solution for the prevention and eradication of paratyphoid fever, it is essential to improve sanitation such as the provision of safe water and food as well as enhanced public health awareness.
Acknowledgements
We thank University of Malaya for facilities and support. This research is financially supported by a University of Malaya High Impact Research Grant—Molecular Genetics (reference no.UM.C/625/1HIR/MOHE/-02 [A000002-5000 1]) and from MOSTI (Molecular Diagnostics of Bacterial pathogens, GA013-2013).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Fangtham M, Wilde H. Emergence of Salmonella Paratyphi A as a major cause of enteric fever: Need for early detection, preventive measures, and effective vaccines. J Travel Med. 2008;15:344-50 doi: 10.1111/j.1708-8305.2008.00237.x
2. Health Protection Agency. Investigation of Faecal Specimens for Enteric Pathogens. UK Standards for Microbiology Investigations. 2013;8(B):30
3. Ochiai RL, Wang XY, von Seidlein L, Yang J, Bhutta ZA, Bhattacharya SK. et al. Salmonella paratyphi A rates, Asia. Emerg Infect Dis. 2005;11:1764-6
4. Girard MP, Steele D, Chaignat CL, Kieny MP. A review of vaccine research and development: human enteric infections. Vaccine. 2006;24:2732-50 doi: 10.1016/j.vaccine.2005.10.014
5. Crump JA, Luby SP, Mintz ED. The global burden of typhoid fever. B World Health Organ. 2004;82:346-53
6. Connor BA, Schwartz E. Typhoid and paratyphoid fever in travellers. The Lancet infectious diseases. 2005;5:623-8 doi:10.1016/S1473-3099(05)70239-5
7. Wilson GS, Miles AA. Principles of Bacteriology and Immunity; Fifth Edition (Vol 1). Great Britain: Edward Arnold (Publishing) Ltd. 1964
8. Thomas HL, Addiman S, Mellanby A. Evaluation of the effectiveness and efficiency of the public health management of cases of infection due to Salmonella typhi/paratyphi in North East London. Public health. 2006;120:1188-93 doi:10.1016/j.puhe.2006.06.013
9. Vollaard AM, Ali S, Widjaja S, Asten HA, Visser LG, Surjadi C. et al. Identification of typhoid fever and paratyphoid fever cases at presentation in outpatient clinics in Jakarta, Indonesia. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2005;99:440-50 doi:10.1016/j.trstmh.2004.09.012
10. Riyaz-Ul-Hassan S, Verma V, Qazi GN. Rapid detection of Salmonella by polymerase chain reaction. Molecular and cellular probes. 2004;18:333-9 doi:10.1016/j.mcp.2004.05.003
11. Parry CM, Hien TT, Dougan G, White NJ, Farrar JJ. Typhoid fever. The New England journal of medicine. 2002;347:1770-82 doi:10.1056/NEJMra020201
12. Gal-Mor O, Suez J, Elhadad D, Porwollik S, Leshem E, Valinsky L. et al. Molecular and cellular characterization of a Salmonella enterica serovar Paratyphi a outbreak strain and the human immune response to infection. Clinical and vaccine immunology: CVI. 2012;19:146-56 doi:10.1128/CVI.05468-11
13. Lolekha S. Salmonella carrier: its evolution and treatment. Sotheast Asian J Trop Med Publ Health. 1995;26:77-9
14. Pegues DA, Michael EO, Miller SI. Salmonella, Including Salmonella Typhi. In Blaser MJ, Smith PD, Ravdin JI, Greenberg HB, Guerrant RL, ed. Infections of the Gastrointestinal Tract. Philadelphia: Lippincott Williams and Wilkins. 2002:669-7
15. Stock I, Wiedemann B. Natural antibiotic susceptibility of Salmonella enterica strains. Int J Antimicrob Ag. 2000;16:211-7 doi: 10.1016/S0924-8579(00)00204-1
16. Edwards PR, Ewing WH. Identification of Enterobacteriaceae; Third Edition. Atlanta, Georgia: Burgess Publishing Company. 1972
17. McClelland M, Sanderson KE, Clifton SW, Latreille P, Porwollik S, Sabo A. et al. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat Genet. 2004;36:1268-74 doi: 10.1038/Ng1470
18. Didelot X, Bowden R, Street T, Golubchik T, Spencer C, McVean G. et al. Recombination and Population Structure in Salmonella enterica. Plos Genet. 2011 7. doi: 10.1371/journal.pgen.1002191
19. Holt KE, Thomson NR, Wain J, Phan MD, Nair S, Hasan R. et al. Multidrug-resistant Salmonella enterica serovar Paratyphi A harbors IncHI1 plasmids similar to those found in serovar Typhi. J Bacteriol. 2007;189:4257-64 doi: 10.1128/Jb.00232-07
20. Mandal S, Mandal MD, Pal NK. Antibiotic resistance of Salmonella enterica serovar Paratyphi A in India: emerging and reemerging problem. Journal of postgraduate medicine. 2006;52:163-6
21. Panigrahi D, Chugh TD, West PWJ, Dimitrov TZ, Groover S, Mehta G. Antimicrobial susceptibility, phage typing and plasmid profile of Salmonella enterica serotype paratyphi a strains isolated in Kuwait. Med Prin Pract. 2003;12:252-5 doi: 10.1159/000072293
22. Huang H, Li J, Yang XL, Wang YG, Wang YP, Tao JS. et al. Sequence Analysis of the Plasmid pGY1 Harbored in Salmonella enterica Serovar Paratyphi A. Biochem Genet. 2009;47:191-7 doi: 10.1007/s10528-008-9216-0
23. Liang WL, Zhao YB, Chen CX, Cui XY, Yu J, Xiao JF. et al. Pan-Genomic Analysis Provides Insights into the Genomic Variation and Evolution of Salmonella Paratyphi A. Plos One. 2012 7. doi: 10.1371/journal.pone.0045346
24. Kumar R, Sazawal S, Sinha A, Sood S, Bhan MK. Typhoid fever: contemporary issues as related to the disease in India. Round table conference series on water borne diseases. 12th ed. Ranbaxy Science Foundation, New Delhi. 1997;2:31-6
25. Nair S, Unnikrishnan M, Turner K, Parija SC, Churcher C, Wain J. et al. Molecular analysis of fluoroquinolone-resistant Salmonella Paratyphi A isolate, India. Emerg Infect Dis. 2006;12:489-91 doi:10.3201/eid1205.050560
26. Kapil A, Sood S, Reddaiah VP, Das B, Seth P. Paratyphoid fever due to Salmonella enterica serotype Paratyphi A. Emerg Infect Dis. 1997;3:407. doi:10.3201/eid0303.970325
27. Sood S, Kapil A, Dash N, Das BK, Goel V, Seth P. Paratyphoid fever in India: An emerging problem. Emerg Infect Dis. 1999;5:483-4
28. Mohanty S, Renuka K, Sood S, Das BK, Kapil A. Antibiogram pattern and seasonality of Salmonella serotypes in a North Indian tertiary care hospital. Epidemiol Infect. 2006;134:961-6 doi: 10.1017/S0950268805005844
29. Capoor MR, Nair D, Deb M, Aggarwal P. Enteric fever perspective in India: emergence of high-level ciprofloxacin resistance and rising MIC to cephalosporins. J Med Microbiol. 2007;56:1131-2 doi: 10.1099/jmm-0.47170-0
30. Lathi N, Sudarsana J, Pushpa KK. Epidemic of Salmonella enterica serotype Paratyphi A in Calicut. Calicut Med J. 2004;2:e2
31. Gupta V, Kaur J, Chander J. An increase in enteric fever cases due to Salmonella Paratyphi A in & around Chandigarh. Indian J Med Res. 2009;129:95-8
32. Rodrigues C, Shenai S, Mehta A. Enteric fever in Mumbai, India: The good news and the bad news. Clin Infect Dis. 2003;36:535 doi: 10.1086/367644
33. Verma S, Thakur S, Kanga A, Singh G, Gupta P. Emerging Salmonella Paratyphi A enteric fever and changing trends in antimicrobial resistance pattern of Salmonella in Shimla. Indian journal of medical microbiology. 2010;28:51-3 doi: 10.4103/0255-0857.58730
34. Tankhiwale SS, Agrawal G, Jalgaonkar SV. An unusually high occurrence of Salmonella enterica serotype Paratyphi A in patients with enteric fever. Indian J Med Res. 2003;117:10-2
35. Mendiratta DK, Deotale V, Thamke D, Narang R, Narang P. Enteric fever due to S. Paratyphi A-an emerging Problem. Indian J Med Microbiol. 2004;22:196
36. Joshi S, Amarnath SK. Fluoroquinolone resistance in Salmonella typhi and S. paratyphi An in Bangalore, India. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2007;101:308-10 doi:10.1016/j.trstmh.2006.05.009
37. Indian Network for Surveillance of Antimicrobial Resistance Group. Antibiogram of S. enterica serovar Typhi and S. enterica serovar Paratyphi A: a multi-centre study from India. WHO South-East Asia J Public Health. 2012;1:182-8
38. Butt T, Ahmad RN, Salman M, Kazmi SY. Changing trends in drug resistance among typhoid salmonellae in Rawalpindi, Pakistan. Eastern Mediterranean health journal = La revue de sante de la Mediterranee orientale = al-Majallah al-sihhiyah li-sharq al-mutawassit. 2005;11:1038-44
39. Khan MI, Soofi SB, Ochiai RL, Khan MJ, Sahito SM, Habib MA. et al. Epidemiology, clinical presentation, and patterns of drug resistance of Salmonella Typhi in Karachi, Pakistan. J Infect Dev Countr. 2012;6:704-14
40. Murdoch DR, Woods CW, Zimmerman MD, Dull PM, Belbase RH, Keenan AJ. et al. The etiology of febrile illness in adults presenting to Patan hospital in Kathmandu, Nepal. The American journal of tropical medicine and hygiene. 2004;70:670-5
41. Woods CW, Murdoch DR, Zimmerman MD, Glover WA, Basnyat B, Wolf L. et al. Emergence of Salmonella enterica serotype Paratyphi A as a major cause of enteric fever in Kathmandu, Nepal. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2006;100:1063-7 doi: 10.1016/j.trstmh.2005.12.011
42. Shirakawa T, Acharya B, Kinoshita S, Kumagai S, Gotoh A, Kawabata M. Decreased susceptibility to fluoroquinolones and gyrA gene mutation in the Salmonella enterica serovar Typhi and Paratyphi A isolated in Katmandu, Nepal, in 2003. Diagn Micr Infec Dis. 2006;54:299-303 doi: 10.1016/j.diagmicrobio.2005.10.016
43. Maskey AP, Basnyat B, Thwaites GE, Campbell JI, Farrar JJ, Zimmerman MD. Emerging trends in enteric fever in Nepal: 9124 cases confirmed by blood culture 1993-2003. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2008;102:91-5 doi:10.1016/j.trstmh.2007.10.003
44. Karkey A, Arjyal A, Anders KL, Boni MF, Dongol S, Koirala S. et al. The Burden and Characteristics of Enteric Fever at a Healthcare Facility in a Densely Populated Area of Kathmandu. Plos One. 2010 5. doi: 10.1371/journal.pone.0013988
45. Pokharel BM, Koirala J, Dahal RK, Mishra SK, Khadga PK, Tuladhar NR. Multidrug-resistant and extended-spectrum beta-lactamase (ESBL)-producing Salmonella enterica (serotypes Typhi and Paratyphi A) from blood isolates in Nepal: surveillance of resistance and a search for newer alternatives. Int J Infect Dis. 2006;10:434-8 doi: 10.1016/j.ijid.2006.07.001
46. Naheed A, Ram PK, Brooks WA, Hossain MA, Parsons MB, Talukder KA. et al. Burden of typhoid and paratyphoid fever in a densely populated urban community, Dhaka, Bangladesh. Int J Infect Dis. 2010;14(Suppl 3):e93-9 doi:10.1016/j.ijid.2009.11.023
47. Sheikh A, Charles RC, Rollins SM, Harris JB, Bhuiyan MS, Khanam F. et al. Analysis of Salmonella enterica serotype paratyphi A gene expression in the blood of bacteremic patients in Bangladesh. PLoS neglected tropical diseases. 2010;4:e908. doi:10.1371/journal.pntd.0000908
48. Yang J. Enteric Fever in South China: Guangxi Province. J Infect Dev Countr. 2008;2:283-8
49. Wu WY, Wang H, Lu J, Wu JS, Chen MJ, Xu YC. et al. Genetic Diversity of Salmonella enterica serovar Typhi and Paratyphi in Shenzhen, China from 2002 through 2007. BMC Microbiol. 2010 10
50. Lam T, Mak D, Wong C. Review of notifiable diseases in Hong Kong, 2010. Public Health Epidemiology Bulletin. 2011;20:31-46
51. Goh KT. An outbreak of paratyphoid A in Singapore: clinical and epidemiological studies. Southeast Asian J Trop Med Public Health. 1981;12:55-62
52. Teoh YL, Goh KT, Neo KS, Yeo M. A nationwide outbreak of coconut-associated paratyphoid a fever in Singapore. Annals of the Academy of Medicine, Singapore. 1997;26:544-8
53. Ty AU, Ang GY, Ang LW, James L, Goh KT. Changing Epidemiology of Enteric Fevers in Singapore. Ann Acad Med Singapore. 2010;39:889-96
54. Jegathesan M. Salmonella serotypes isolated from man in Malaysia over the 10-year period 1973-1982. The Journal of hygiene. 1984;92:395-9
55. Yasin RM, Jegathesan MM, Tiew CC. Salmonella serotypes isolated in Malaysia over the ten-year period 1983-1992. Asia-Pacific journal of public health / Asia-Pacific Academic Consortium for Public Health. 1996;9:1-5
56. Commons RJ, McBryde E, Valcanis M. et al. Twenty-six years of enteric fever in Australia: an epidemiological analysis of antibiotic resistance. Med J Aust. 2012;196:332-6
57. Blackstock SJ, Sheppeard VK, Paterson JM, Ralph AP. Typhoid and paratyphoid fever in Western Sydney Local Health District, NSW, January-June 2011. New South Wales public health bulletin. 2012;23:148-52 doi:10.1071/NB11041
58. Threlfall EJ, Fisher IST, Berghold C, Gerner-Smidt P, Tschape H, Cormican M. et al. Trends in antimicrobial drug resistance in Salmonella enterica serotypes Typhi and Paratyphi A isolated in Europe, 1999-2001. Int J Antimicrob Ag. 2003;22:487-91 doi: 10.1016/S0924-8579(03)00262-0
59. Health Protection Agency. Foreign travel-associated illness in England, Wales and Northern Ireland: 2007 report. London: Health Protection Agency. 2007
60. Health Protection Agency. Migrant Health: Infectious diseases in non-UK born populations in the United Kingdom. An update to the baseline report-2011.London: Health Protection Agency. 2011
61. Health Protection Agency. Enteric fever surveillance quarterly report (England, Wales and Northern Ireland) - first quarter 2012. HPR Weekly [serial online]. 2012;6(19):enteric
62. Health Protection Agency. Enteric fever surveillance quarterly report (England, Wales and Northern Ireland) - second quarter 2012. HPR Weekly [serial online]. 2012;6(32):enteric
63. Health Protection Agency. Enteric fever surveillance quarterly report (England, Wales and Northern Ireland) - third quarter 2012. HPR Weekly [serial online]. 2012;6(45):enteric
64. Health Protection Agency. Enteric fever surveillance quarterly report (England, Wales and Northern Ireland) - fourth quarter 2012. HPR Weekly [serial online]. 2013;7(6):enteric
65. Caumes E, Ehya N, Nguyen J, Bricaire F. Typhoid and paratyphoid fever: A 10-year retrospective study of 41 cases in a Parisian hospital. J Travel Med. 2001;8:293-7
66. Walia M, Gaind R, Paul P, Mehta R, Aggarwal P, Kalaivani M. Age-related clinical and microbiological characteristics of enteric fever in India. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2006;100:942-8 doi: 10.1016/j.trstmh.2006.02.015
67. Khuribulos N. Enteric Fevers in Children - the Importance of Age in the Varying Clinical Picture. Clin Pediatr. 1981;20:448-52 doi: 10.1177/000992288102000704
68. Joshi S, Wattal C, Sharma A, Prasad KJ. Mixed Salmonella infection - a case report. Indian journal of medical microbiology. 2002;20:113-4
69. Humphries RM, Yeganeh N, Ward KW, Lewinski MA, Ching N. Enteric fever in a 6-year-old traveler caused by Salmonella enterica serotypes Typhi and Paratyphi A: laboratory detection strategies and treatment options. Journal of clinical microbiology. 2011;49:452-4 doi:10.1128/JCM.01316-10
70. Walker RA, Skinner JA, Ward LR, Threlfall EJ. LightCycler gyrA mutation assay (GAMA) identifies heterogeneity in GyrA in Salmonella enterica serotypes Typhi and Paratyphi A with decreased susceptibility to ciprofloxacin. Int J Antimicrob Agents. 2003;22:622-5
71. Chandel DS, Chaudhry R, Dhawan B, Pandey A, Dey AB. Drug-resistant Salmonella enterica serotype paratyphi A in India. Emerg Infect Dis. 2000;6:420-1 doi:10.3201/eid0604.000420
72. Bhattacharya SS, Das U, Choudhury BK. Occurrence & antibiogram of Salmonella Typhi & S. Paratyphi A isolated from Rourkela, Orissa. Indian J Med Res. 2011;133:431-3
73. Dimitrov T, Udo EE, Albaksami O, Al-Shehab S, Kilani A, Shehab M. et al. Clinical and microbiological investigations of typhoid fever in an infectious disease hospital in Kuwait. J Med Microbiol. 2007;56:538-44 doi:10.1099/jmm.0.46814-0
74. Zaki SA, Karande S. Multidrug-resistant typhoid fever: a review. J Infect Dev Ctries. 2011;5:324-37
75. Parry CM, Ho VA, Phuong le T, Bay PV, Lanh MN, Tung le T. et al. Randomized controlled comparison of ofloxacin, azithromycin, and an ofloxacin-azithromycin combination for treatment of multidrug-resistant and nalidixic acid-resistant typhoid fever. Antimicrobial agents and chemotherapy. 2007;51:819-25 doi:10.1128/AAC.00447-06
76. Crump JA, Mintz ED. Global Trends in Typhoid and Paratyphoid Fever. Clin Infect Dis. 2010;50:241-6 doi: 10.1086/6495
77. Dolecek C, Tran TP, Nguyen NR, Le TP, Ha V, Phung QT. et al. A multi-center randomised controlled trial of gatifloxacin versus azithromycin for the treatment of uncomplicated typhoid fever in children and adults in Vietnam. Plos One. 2008;3:e2188. doi:10.1371/journal.pone.0002188
78. Trivedi NA, Shah PC. A meta-analysis comparing the safety and efficacy of azithromycin over the alternate drugs used for treatment of uncomplicated enteric fever. Journal of postgraduate medicine. 2012;58:112-8 doi: 10.4103/0022-3859.97172
79. Aggarwal A, Ghosh A, Gomber S, Mitra M, Parikh AO. Efficacy and safety of azithromycin for uncomplicated typhoid fever: an open label non-comparative study. Indian pediatrics. 2011;48:553-6
80. Sjolund-Karlsson M, Joyce K, Blickenstaff K, Ball T, Haro J, Medalla FM. et al. Antimicrobial susceptibility to azithromycin among Salmonella enterica isolates from the United States. Antimicrobial agents and chemotherapy. 2011;55:3985-9 doi:10.1128/AAC.00590-11
81. Molloy A, Nair S, Cooke FJ, Wain J, Farrington M, Lehner PJ. et al. First report of Salmonella enterica serotype paratyphi A azithromycin resistance leading to treatment failure. Journal of clinical microbiology. 2010;48:4655-7 doi:10.1128/JCM.00648-10
82. Pancharoen C, Thisyakorn C, Thisyakorn U. Endocarditis and pericarditis caused by Salmonella paratyphi A: two case reports and review of the literature. The Southeast Asian journal of tropical medicine and public health. 2002;33:161-3
83. Sanchez-Guerrero J, Alarcon-Segovia D. Salmonella pericarditis with tamponade in systemic lupus erythematosus. British journal of rheumatology. 1990;29:69-71
84. Gupta S, Singh B, Kapoor H, Minocha SK, Jain AK. Ventricular septal defect with endocarditis caused by Salmonella Paratyphi A. Trop Doct. 1994;24:40
85. Johnson DH, Rosenthal A, Nadas AS. Bacterial endocarditis in children under 2 years of age. American journal of diseases of children. 1975;129:183-6
86. Chaudhry R, Chandel DS, Pandey A, Dhawan B, Gulati V, Sood R. Utility of PCR in diagnosing complicated cases of unusual clinical manifestations of Salmonella enterica var. Paratyphi A. The American journal of medicine. 2005;118:799-800 doi:10.1016/j.amjmed.2004.12.030
87. Hu L, Kopecko DJ. Salmonella Typhi and Paratyphi. In Sussman M, ed. Molecular Medical Microbiology 2. Carlifornia: Academic Press. 2002:1365-91
88. Kudalkar D, Thermidor M, Cunha BA. Salmonella paratyphi A enteric fever mimicking viral meningitis. Heart & lung: the journal of critical care. 2004;33:414-6
89. Jeans AR, McKendrick MW. Salmonella Paratyphi A liver abscess--secondary infection of an amoebic liver abscess? Travel medicine and infectious disease. 2007;5:144-6 doi:10.1016/j.tmaid.2006.01.004
90. Wain J, Diep TS, Ho VA, Walsh AM, Nguyen TT, Parry CM. et al. Quantitation of bacteria in blood of typhoid fever patients and relationship between counts and clinical features, transmissibility, and antibiotic resistance. Journal of clinical microbiology. 1998;36:1683-7
91. Ekpo P, Sarasombath S. Expression of Salmonella Paratyphi B and Salmonella Paratyphi C flagellin/glutathione S-transferase fusion protein. Sotheast Asian J Trop Med Publ Health. 1999;26(Suppl2):S192-S194
92. Parry CM, Hoa NTT, Diep TS, Wain J, Chinh NT, Vinh H. et al. Value of a single-tube Widal test in diagnosis of typhoid fever in Vietnam. Journal of clinical microbiology. 1999;37:2882-6
93. Willke A, Ergonul O, Bayar B. Widal test in diagnosis of typhoid fever in Turkey. Clinical and diagnostic laboratory immunology. 2002;9:938-41
94. Tam FCH, Wang ML, Dong BQ, Leung DTM, Ma CH, Lim PL. New rapid test for paratyphoid a fever: usefulness, cross-detection, and solution. Diagn Micr Infec Dis. 2008;62:142-50 doi: 10.1016/j.diagmicrobio.2008.07.002
95. Tracz DM, Tabor H, Jerome M, Ng LK, Gilmour MW. Genetic determinants and polymorphisms specific for human-adapted serovars of Salmonella enterica that cause enteric fever. Journal of clinical microbiology. 2006;44:2007-18 doi: 10.1128/Jcm.02630-05
96. Nissen MD, Sloots TP. Rapid diagnosis in pediatric infectious diseases: the past, the present and the future. Pediatr Infect Dis J. 2002;21:605-12 discussion 13-4
97. Hirose K, Itoh K, Nakajima H, Kurazono T, Yamaguchi M, Moriya K. et al. Selective amplification of tyv (rfbE), prt (rfbS), viaB, and fliC genes by multiplex PCR for identification of Salmonella enterica serovars Typhi and Paratyphi A. Journal of clinical microbiology. 2002;40:633-6
98. Ou HY, Teh CSJ, Thong KL, Ahmad N, Deng ZX, Barer MR. et al. Translational genomics to develop a Salmonella enterica serovar Paratyphi A multiplex polymerase chain reaction assay. J Mol Diagn. 2007;9:624-30 doi: 10.2353/jmoldx.2007.070064
99. Teh CSJ, Chua KH, Puthucheary SD, Thong KL. Further evaluation of a multiplex PCR for differentiation of Salmonella Paratyphi A from other Salmonellae. Jpn J Infect Dis. 2008;61:313-4
100. Nga TV, Karkey A, Dongol S, Thuy HN, Dunstan S, Holt K. et al. The sensitivity of real-time PCR amplification targeting invasive Salmonella serovars in biological specimens. Bmc Infect Dis. 2010;10:125. doi:10.1186/1471-2334-10-125
101. Arya SC, Sharma KB. Urgent need for effective vaccine against Salmonella paratyphi A, B and C. Vaccine. 1995;13:1727-8 doi: 10.1016/0264-410x(96)81276-X
102. Konadu EY, Lin FYC, Ho VA, Thuy NTT, Bay PV, Thanh TC. et al. Phase 1 and phase 2 studies of Salmonella enterica serovar Paratyphi A O-specific polysaccharide-tetanus toroid conjugates in adults, teenagers, and 2-to 4-year-old children in Vietnam. Infect Immun. 2000;68:1529-34 doi: 10.1128/Iai.68.3.1529-1534.2000
103. Roland KL, Tinge SA, Kochi SK, Thomas LJ, Killeen KP. Reactogenicity and immunogenicity of live attenuated Salmonella enterica serovar Paratyphi A enteric fever vaccine candidates. Vaccine. 2010;28:3679-87 doi:10.1016/j.vaccine.2010.03.019
104. Gat O, Galen JE, Tennant S, Simon R, Blackwelder WC, Silverman DJ. et al. Cell-associated flagella enhance the protection conferred by mucosally-administered attenuated Salmonella Paratyphi A vaccines. PLoS neglected tropical diseases. 2011;5:e1373. doi:10.1371/journal.pntd.0001373
105. Micoli F, Rondini S, Gavini M, Lanzilao L, Medaglini D, Saul A. et al. O:2-CRM(197) conjugates against Salmonella Paratyphi A. Plos One. 2012;7:e47039. doi:10.1371/journal.pone.0047039
Author contact
![]() Corresponding author: Kwai Lin Thong, PhD. Professor, Institute of Biological Sciences, Faculty of Science, University of Malaya, 50603 Kuala Lumpur. Tel: +603-79674436; Fax: +603-79675908 Email: thongkledu.my.
Corresponding author: Kwai Lin Thong, PhD. Professor, Institute of Biological Sciences, Faculty of Science, University of Malaya, 50603 Kuala Lumpur. Tel: +603-79674436; Fax: +603-79675908 Email: thongkledu.my.

 Global reach, higher impact
Global reach, higher impact