Impact Factor
ISSN: 1449-1907
Int J Med Sci 2014; 11(7):674-679. doi:10.7150/ijms.6962 This issue Cite
Research Paper
Cytokine Gene Polymorphisms Associate with Microbiogical Agents in Periodontal Disease: Our Experience
1. Department of Basic Medical Sciences, Neurosciences and Sense Organs, University of Bari “Aldo Moro”, Piazza Giulio Cesare 11, 70124, Bari, Italy.
2. Department of Interdisciplinary Medicine, University of Bari “Aldo Moro”, Piazza Giulio Cesare 11, 70124, Bari, Italy.
Received 2013-6-20; Accepted 2014-3-11; Published 2014-5-1
Abstract
Periodontics has evolved from a simplistic model to a more complex interplay between infection and host response. Genetic factors have been a new addition to the list of risk factors for periodontal diseases. The processes leading to destruction and regeneration of the destroyed tissues are of great interest to both researchers and clinicians.
The selective susceptibility of subjects for periodontitis has remained an enigma and wide varieties of risk factors have been implicated for the manifestation and progression of periodontitis. Emerging pathway models suggest that gene-environment interactions are etiologically important in disease pathogenesis. The current practical utility of genetic knowledge in periodontitis is limited.
Allelic variants at multiple gene loci probably influence periodontitis susceptibility. The pro-inflammatory cytokine interleukin-1 (IL-1) is a key modulator of host responses to microbial infection and a major modulator of extracellular matrix catabolism and bone resorption, and polymorphisms in the IL-1 gene cluster have been associated with an increased risk of developing severe adult periodontitis.
The aim of this study was to test if polymorphisms of genes of IL-1α+4845 and IL-1β +3954 were linked with periodontitis, in a case-control study population, delimited to a specific geographic area, in association with microbiological findings.
The polymorphisms observed in IL-1α+4845 and IL-1β+3954 single nucleotide polymorphisms (SNPs), was significantly different among the study groups (healthy controls, mild, moderate and severe periodontitis with p<0.05, d.f.=1.
We found a significant correlation between the severe form of periodontitis and the presence of composite genotype (p < 0.05, d.f.=1, calculated among healthy vs. severe). Furthermore a statistically significant association between the presence of bacteria and periodontitis was detected (p<0.05, d.f.=1). In the current investigation findings were concordant with literature observations.
Keywords: Periodontal disease, Single nucleotide polymorphisms, Interleukin-1, Genetic factors, Oral Microbiology.
Introduction
Inflammation is clearly a central component of many chronic diseases. Cytokines are regulatory low-weight proteins acting as mediators into genesis and control of immune and inflammatory response.
Interleukin-1 (IL-1) is a proinflammatory cytokine and includes two different but functionally similar molecules: IL-1α e IL-1β, genes which are all located on the long arm of chromosome 2 (region 2q13-14). [1-3]
IL-1 play an important role in immune regulation and inflammatory processes by inducing expression of many effector proteins, e.g. cytokines/chemokines, nitric oxide synthetase (NOS) and matrix metalloproteinases (MMPs) [1,2].
IL-1α is mainly produced by keratinocytes of junctional epithelium or pocket epithelium, is a mediator of local inflammation and regulates intracellular events. [1]
IL-1β is primarily produced by activated macrophages and fibroblasts that release this extracellular protein. [1]
IL-1 upregulates complement system, Fc-receptor on polymorphonuclear neutrophils and monocytes, and the expression of adhesion molecules upon fibroblast and leukocytes. [1, 4]
It induces the expression of homing receptors into the extracellular matrix for leukocytes and bone resorption by stimulating the proliferation and differentiation of osteoclast progenitors and activating formed osteoclast indirectly. [5]
It increases the production of MMPs, prostaglandins and IL-1 itself by macrophages, fibroblast and polymorphonuclear neutrophils. [5,6]
It upregulates the expression of major histocompatibility complex molecules (MHC) upon B and T cells facilitating their activation, clonal expansion and immunoglobulin production. [6]
Excessive and/or dysregulated activity of these mediators is associated with tissue destruction and therefore the synthesis, secretion and biological activity of IL-1 cytokines have been identified as therapeutic targets for common inflammatory disorders such as rheumatoid arthritis (RA), diabetes and periodontal disease [7-9].
Increase of IL-1 gingival concentration in subjects with periodontal disease and its reduction after periodontal treatment was demonstrated. [10,11]
Periodontal disease (PD) is characterized by loss of connective tissues within the periodontium and destruction of (alveolar) bone support. [12-15]
The most prevalent form of PD is chronic periodontitis, which is an inflammatory disease initiated by pathogenic bacteria in the subgingival plaque biofilm. [12]
Furthermore, a prominent, localized inflammatory cell infiltrate involving neutrophils, monocytes and both T and B lymphocytes is another characteristic these cells are further sources of MMPs and the cytokines that regulate them.[5,6]
Single nucleotide polymorphisms (SNPs) into the IL-1 promoter region could modulate this cytokine function that acts a fundamental role into the pathogenesis of PD, by affecting the susceptibility and severity of the affection. [16-18]
The composite IL-1 genotype, known as periodontitis-associated genotype (PAG), obtained by the combination of the two rare alleles at the above mentioned SNPs, has been extensively investigated with respect to the form of periodontal disease (chronic vs aggressive), smoke, ethnic group, oral microbial pathogens present in the plaque biofilm and other periodontal clinical parameters related to inflammatory status or to the progression of the disease. [16,17]
The aim of this study was to test if polymorphisms of genes of IL-1α+4845 and IL-1β +3954 were linked with periodontitis, in a case-control study population, delimited to a specific geographic area, in association with microbiological findings.
Patients, Materials and Methods
All subjects were chosen among current and permanent residents of Bari area (Italy).
Apulia (Italian: Puglia) is a Region in Southern Italy. The Region comprises 19,345 square kilometers (7,469 sq mi), and its Population is about 4.1 million Density 210/km2 (540/sq mi). Its capital city is Bari.
Population proportionate technique was employed for sample size estimation. The city is divided in 10 quarters. The average age of Bari residents is 42 compared to the Italian average of 42. The present study includes only resident form 4 generations in the quarter of Old Town that is the one with less heterogeneity of population. According to population census 2011, city itself has a population of about 320.000, over 116 km2; the Old Town in an area of around 0,4 square Kilometers, have a population of around 6000 people, of which the 26-56 old adults constitute 33%. With 95% confidence level, the estimated sample size was 99.
This work was conducted according to the recommendations of the Declaration of Helsinki; all patients received informed consent.
The study included as described in Table 1 and Table 2, 195 unrelated patients, recruited only from private dental practice.
All subjects were in good general health conditions.
To identify suitable participants, patient referral records, including a full-mouth series of peri-apical radiographs, were screened.
Complete examinations of soft and hard oral tissues were performed on all patients.
Exclusion criteria from the study were: diseases of the oral hard or soft tissues except caries and periodontal disease; presence/use of orthodontic appliances; need of pre-medication for dental treatment; use of antibiotics for the preceding 6-month period; pregnancy or lactation; inability or unwillingness to sign informed consent; history of diabetes; HIV positive; immunosuppressive chemotherapy; history of any disease known to severely compromise immune function.
Gender characteristic and data of Patients with Periodontal Disease and Control Subjects.
| Cases | Controls | |||
|---|---|---|---|---|
| Age (years) | 54.25 (mean) | 32 to 56 (range) | 48.15 (mean) | 26 to 56 (range) |
| Male | 52 | 52.5% | 49 | 51% |
| Female | 47 | 47.5% | 47 | 49% |
| Smokers | 33 | 33.3% | 24 | 25% |
| No smokers | 66 | 66.7% | 72 | 75% |
Cases population divided by periodontitis severity.
| Subjects | % | |
|---|---|---|
| Mild periodontitis (Group B) | 40 | 40.4 |
| Moderate periodontitis (Group C) | 39 | 39.4 |
| Severe periodontitis (Group D) | 20 | 20.2 |
| Total | 99 | 100 |
No subject with localized periodontitis or acute necrotizing ulcerative gingivitis was included in the study.
All patients were interviewed and smoking habits were recorded. They were subjected to a clinical examination of the periodontal tissues.
The parameters investigated were: plaque accumulation (0/1/2), overt gingivitis (0/1), bleeding on probing (0/1), and suppuration (0/1).
A manual periodontal probe was used to assess the attachment loss which was measured at six sites per tooth at all teeth excluding third molars.
Regarding these parameters, the cases were divided into four groups:
(A) Healthy controls patients;
(B) With mild periodontitis (plaque accumulation, overt gingivitis, attachment loss ≤ 4 mm with or without bleeding);
(C) Moderate periodontitis (bleeding and/or attachment loss between 4-6 mm);
(D) Severe periodontitis (bleeding or suppuration and attachment loss > 6 mm).
Periodontally intact subjects were qualified as controls if they displayed: no sites with probing depth exceeding 3 mm and no more than 4 sites with bleeding on probing.
Our study recruited only Italian Caucasian subjects, and none of them had a history of diabetes mellitus or current manifestation of systemic diseases, which correlates with destructive periodontal disease.
Radiographic evaluation included Orthopantomography and 21 intraoral radiographs.
All data from clinical and radiographic evaluation were collected into a dedicated periodontal folder.
The periodontal probe (PCP15 -HuFriedy, Chicago, IL, USA) was inserted parallel to the vertical axis of the tooth and 'walked' circumferentially clockwise around each surface of the tooth, to detect the area of deepest penetration [19].
Genomic Analysis
Analysis of genetic polymorphisms
Analysis of blood samples was done in a blinded fashion.
Five milliliters of venous blood was drawn from the antecubital vein and transferred to a vacutainer containing ethylenediaminetetraacetic acid (EDTA; 3%) to prevent coagulation and stored at -20°C.
Genomic DNA was extracted by the salting out method [20]: DNA was aliquoted in distilled water and the stocks were stored at 4°C; DNA integrity was also tested, by running genomic DNA through 0.8% agarose gel, and visualized with ethidium bromide.
We tested the following variants by using primer concentrations, optimized thermocycling conditions and gel compositions as already published in the cited literature: IL-1A+4845 [21], IL-1B+3954 [22].
The genomic DNA regions were amplified using PCR primers annealing to positions flanking the polymorphisms. IL-1α +4845 G>T(rs17561), and IL-1β +3954 C>T (rs1143634) with the primer sequences and PCR conditions given in Table 3, using a Q-PCR thermocycler.
The primer sequences and the genotyping methods for IL-1α and IL-1β polymorphisms.
| LOCUS AND PRIMER SEQUENCES | ANNEALING TEMPERATURE (°C) | PCR AMPLICON SIZE (BP) |
|---|---|---|
| IL-1α+4845 | 56 | 229 |
| F:5'ATGGTTTTAGAAATCATCAAGCCTAGGGCA3' | ||
| R:5'AATGAAAGGAGGGGAGGATGACAGAAATT3' | ||
| IL-1β+3954 | 58 | 194 |
| F: 5' CTCAGGTGTCCTCGAAGAAATCAA 3' | ||
| R: 5' GCTTTTTTGCTGTGAGTCCCG 3' |
For a more accurate identification of SNPs, we purified 2-3 µl of some PCR products with Exonuclease I and Alcaline Phosphatase USB (Amersham Pharmacia Biotech, Cologno Monzese, Italy) enzymes; then they were sent for sequencing with a DNA Analyzer (ABI PRISM 3700 Applied Biosystems, Monza, Italy). For all markers, primers were from MWG Biotech (Firenze, Italy).
DNA Sequencing was performed by modified dideoxy mediated chain-termination method, using the ABI Prism (Applied Biosystems, USA) 3130 genetic analyzer automated DNA sequencer and ABI Prism Big Dye Terminator v3.1 cycle sequencing kit.
Accuracy of the method was confirmed by reanalyzing mutant samples, and representative samples of the genotypes of each polymorphic loci was sequenced to confirm the mutations detected.
MicroDent test
Subgingival microbial species were determined in each plaque sample using a PCR technique (micro-IDent® plus 11 test, Hain Diagnostika Ltd. - Nehren, Germany), according to the manufacturer's recommendations. [23-26]
The micro-IDent® plus 11 test is a fast and reliable DNA hybridization method to identify microbial subgingival species.
During the clinical examination, the periodontal plaque samples (PS) were collected as follows: the supragingival plaque was removed with an ultrasonic device and with gentle rotary brushing from central lower incisors and mandibular second molars (caries-free periodontal sites with the deepest, inflamed periodontal pockets and attachment loss).
The periodontal sites to be sampled were air-dried and isolated with cotton rolls, and two sterile paper points ISO size 40 (Dentsply Maillefer, Ballaigues, Switzerland) were inserted into periodontal pocket [27] after 1 min, the paper points were removed, pooled and frozen immediately at -80 °C in a separate sterile 2-mL cryotube filled with 1 mL of sterile phosphate-buffered saline (PBS, 50 mmol L -1 potassium phosphate, 150 mmol L-1 NaCl, pH 7.2.
For the rapid manual isolation of genomic bacterial DNA from patient specimen we used GenoLyse® kit (Hain Diagnostika Ltd. - Nehren, Germany), according to the manufacturer's recommendations.
After PCR amplification, evaluation of the results was performed with a specific reading card that allows to immediately identifying the microbial species.
Statistical Analysis
Statistical analysis of data was performed using the NCSS8¸ Statistical Software. The Chi-square test was used to compare the genotype distributions between the control and PD groups (d.f.=1). A p-value of < 0.05 was considered significant.
Results
The present study determined the frequencies of the two IL-1 polymorphisms, IL-1α+4845 and IL-1β+3954 in a sample group of 195 individuals.
Sixty-four subjects were positive for IL-1α+4845 genotype (33 %), with a major frequency of positive subjects observed among the group B subjects and the least frequency observed among the healthy subjects. (Table 4)
This difference achieved statistical significance because p < 0.05 at d.f.=1.
Similar trend was noticed for the SNPs of IL-1β+3954 gene, with the frequency progressively increasing from group A through group C, B, to group D, which was significant at p<0.05, d.f.=1. (Table 5)
The combined presence of both the IL-1α and IL-1β composite genotype, dived for severity of disease was studied. (Table 6)
This difference brought out a significant correlation between the severe form of periodontitis and the presence of composite genotype (p < 0.05, d.f.=1, calculated among healthy vs. severe).
Furthermore, microbiological data were available for all the subgingival plaque samples collected from all cases and controls. (Table 7)
As expected a statistically significant association between the presence of bacteria and periodontitis was detected (p<0.05, d.f.=1).
Distribution of IL-1 positive genotype polymorphisms in periodontitis (divided for disease severity) and controls.
| Severe periodontitis | Moderate periodontitis | Mild periodontitis | Controls | Total | |
|---|---|---|---|---|---|
| IL-1α (+) | 7 | 20 | 21 | 116 | 64 |
| IL-1α (-) | 13 | 19 | 19 | 80 | 131 |
| Total | 20 | 39 | 40 | 96 | 195 |
Distribution of IL-1β genotype polymorphisms in periodontitis (divided for disease severity) and controls.
| Severe periodontitis | Moderate periodontitis | Mild periodontitis | Controls | Total | |
|---|---|---|---|---|---|
| IL-1β (+) | 61 | 5 | 6 | 06 | 23 |
| IL-1β (-) | 14 | 134 1 | 34 | 90 | 173 |
| Total | 20 | 39 | 40 | 96 | 195 |
Distribution of IL-1 composite genotype polymorphisms in periodontitis (divided for disease severity) and controls.
| Severe periodontitis | Moderate periodontitis | Mild periodontitis | Controls | Total | |
|---|---|---|---|---|---|
| IL-1α (+)/IL-1β (+) | 3 | 3 | 2 | 2 | 10 |
| IL-1α(+)/IL-1β(-) | 4 | 17 | 19 | 14 | 54 |
| IL-1α(-)/IL-1β(+) | 3 | 2 | 4 | 4 | 13 |
| IL-1α(-)/IL-1β(-) | 10 | 17 | 15 | 76 | 118 |
Pathogens presence in periodontitis (divided for disease severity) and healthy controls.
| Severe periodontitis | Moderate periodontitis | Mild periodontitis | Controls | Total | |
|---|---|---|---|---|---|
| Bacteria (+) | 20 (100%) | 39 (100%) | 40 (100%) | 50 (52%) | 149 |
| Bacteria (-) | 0 (0%) | 0 (0%)0 | 0 (0%) | 46 (48%) | 46 |
| Total | 20 (100%) | 39 (100%) | 40 (100%) | 96 (100%) | 195 |
The most frequent pathogens found in the subgingival plaque of patients positives for the presence of bacteria were Fusobacterium nucleatum (27%), Campylobacter rectus (20%), Eikenella corrodens (20%), Tannerella forsythia (2%), Treponema denticola (6%), Parvimonas micra (7%),, Porphyromonas gingivalis (10%), and Prevotella intermedia (8%). (Figure 1)
When the allele distribution was compared between cases and healthy controls, statistically insignificant values were obtained. (Table 8)
The heterozygous genotypes (GT) and (CT) are more frequent respectively in, IL-1α+4845 and IL-1β+3954, for both cases and controls.
Pathogens found in the subgingival plague.
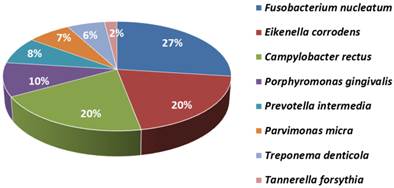
Summary of allele and genotype distribution in cases and healthy controls.
| Locus | Genotype | Allele | P-value | ||||
|---|---|---|---|---|---|---|---|
| IL-1α(+) | GG | GT | TT | G | T | ||
| Cases | 18 | 28 | 2 | 60 | 36 | 0.19 | |
| Controls | 4 | 11 | 1 | 24 | 8 | ||
| IL-1β(+) | CC | CT | TT | C | T | ||
| Cases | 7 | 9 | 1 | 28 | 6 | 0.93 | |
| Controls | 2 | 4 | 0 | 10 | 2 | ||
In the current investigation findings were concordant with literature observations. [21-22].
However, contradictory results were obtained in a recently published data [28] where they have found that for IL-1β+3954 CC genotype was the more frequently genotype identified in cases than in controls.
Discussion
In the present work, we have directed our effort primarily on the study of a homogeneous population from both ethnic and clinical points of examination.
This approach, given the small extent of our region, reduces the possibilities for establishing large sample sizes, but the homogeneous population has fewer problems with population stratification.
A second limitation of the present study is represented by the limited size of case samples.
Current evidence from complex diseases (with large heritable components) indicates that the predicted odds ratios for many genes may be 1.5 or less and, to identify such associations, as a general rule at least 100 cases and controls should be genetically characterized. [29, 30]
The subjects may not be representative of the true disease distribution because of recruitment from a clinical practice.
This phenomenon is known as the "clinicians' illusion," where the identified cases among the subjects are likely to be more numerous and severe than cases identified from a general representative population sample. [31]
The sample is very small as to provide for any generalized conclusions based on the results that apply to the whole population.
Due to the phenomenon of genetic heterogeneity, the periodontal phenotype may be due to several different genotypes. [21-22, 32-33]
The clinical categorization of PD has been different in different studies due to changes in classification and nomenclature.
Since many risk factors contribute to PD, the ideal method of studying genetic relationship would be to start before the disease sets in and following it up through the natural history, i.e., to establish the genotypes required first and then follow it up as the subject is exposed to various predisposing and risk factors.
Also, as recommended, further researches are needed to validate the biologic basis for genetic susceptibility testing, to evaluate the ability of IL-1 genotypes to predict disease initiation and to evaluate the effectiveness of IL-1 genotyping in making diagnostic or treatment intervention strategies, especially in dental “new age” [34] tissue engineering approach [35-40].
Acknowledgements
The Authors express their gratitude towards Dr. Vincenzo Altini and Dr. Giuseppe Zaccaria, for their support in patients' recruitment.
Authors' contributions
DdV supervised and coordinated the research. SS, FI and FP revised it critically for important intellectual content and gave final approval of the version to be published. AB made substantial contributions to conception and design and edited the manuscript. SC assisted the clinical procedures and selected the patients performing all the clinical measurements and the database. GD was responsible for bibliographic research. RM and FMC were responsible for the statistical elaboration and graphics layout of the manuscript.
Competing Interests
The Authors declared that no competing interest exists. None of the funding sources had any role in the design and conduct of the study. All Authors reviewed the paper, and read and approved the final manuscript.
References
1. Dinarello CA. The IL-1 family and inflammatory diseases. Clin Exp Rheumatol. 2002;20:S1-13
2. Dinarello CA. Biologic basis for interleukin-1 in disease. Blood. 1996;87:2095-2147
3. Collins A, Morton NE. Mapping a disease locus by allelic association. Proc Natl Acad Sci USA. 1998;95:1741-1745
4. Chai L, Song YQ. Leung WK. Genetic polymorphism studies in periodontitis and Fcγ receptors. J Periodont Res. 2011;47(3):273-285
5. Gemmell E, Marshall RI, Seymour G. Cytokines and prostaglandins in immune homeostasis and tissue destruction in periodontal disease. Periodontology 2000. 1997;14:112-143
6. Milner JM, Cawston TE. Matrix metalloproteinase knockout studies and the potential use of matrix metalloproteinase inhibitors in the rheumatic diseases. Curr Drug Targets Inflamm Allergy. 2005;4:363-75
7. Kay J, Calabrese L. The role of interleukin-1 in the pathogenesis of rheumatoid arthritis. Rheumatology (Oxford). 2004;43(Suppl. 3):2-9
8. Bartold PM, Marshall RI, Haynes DR. Periodontitis and rheumatoid arthritis: a review. J Periodontol. 2005;76:2066-74
9. Donath MY, Shoelson SE. Type 2 diabetes as an inflammatory disease. Nat Rev Immunol. 2011Feb;11(2):98-107
10. Masada MP, Persson R, Kenney JS, Lee SW, Page RC, Allison AC. Measurement of interleukin-1α and -1β in gingival crevicular fluid: implication for the pathogenesis of periodontal disease. J Periodont Res. 1990;25:156-163
11. Greenstein G, Hart TC. A critical assessment of interleukin 1 (IL-1) genotyping when used in a genetic susceptibility test for severe chronic periodontitis. J Periodontol. 2002;73:231-47
12. Pihlstrom BL, Michalowicz BS, Johnson NW. Periodontal diseases. Lancet. 2005;366:1809-20
13. Ballini A, Tetè S, Scattarella A, Cantore S, Mastrangelo F, Papa F, Nardi GM, Perillo L, Crincoli V, Gherlone E, Grassi FR. The Role Of Anti-Cyclic Citrullinated Peptide Antibody In Periodontal Disease. Int J Immunopathol Pharmacol. 2010;23(2):677-681
14. Mori G, Brunetti G, Colucci S, Oranger A, Ciccolella F, Sardone F, Pignataro P, Mori C, Karapanou V, Ballini A, Mastrangelo F, Tete S, Grassi F.R, Grano M. Osteoblast Apoptosis In Periodontal Disease: Role of TNF-Related Apoptosis-Inducing Ligand. Int J Immunopathol Pharmacol. 2009;22(1):95-103
15. Mori G, Brunetti G, Colucci S, Ciccolella F, Coricciati M, Pignataro P, Oranger A, Ballini A, Farronato D, Mastrangelo F, Tetè S, Grassi FR, Grano M. Alteration of activity and survival of osteoblasts obtained from human periodontitis patients: role of TRAIL. J Biol Regul Homeost Agents. 2007;21(3-4):105-14
16. Kinane DF, Hart TC. Genes and gene polymorphisms associated with periodontal disease. Crit Rev Oral Biol Med. 2003;14:430-449
17. McDevitt MJ, Wang HY, Knobelman C. et al. Interleukin-1 genetic association with periodontitis in clinical practice. J Periodontol. 2000;71:156-163
18. Kornman KS, Crane A, Wang HY, di Giovine FS, Newman MG, Pirk FW. et al. The interleukin-1 genotype as a severity factor in adult periodontal disease. J Clin Periodontol. 1997;34:72-7
19. Carranza FA, Takei HH. Clinical diagnosis. In: (ed.) Newman MG, Takei HH, Klokkevold PR, Carranza F. Carranza's Clinical Periodontology, 10th edn. St. Louis, MO, USA: Saunders, Elsevier. 2006:548-58
20. Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215
21. Mao M, Zeng XT, Ma T, He W, Zhang C, Zhou J. Interleukin-1α -899 (+4845) C→T polymorphism increases the risk of chronic periodontitis: evidence from a meta-analysis of 23 case-control studies. Gene. 2013;532(1):114-9 doi: 10.1016/j.gene.2013.09.043. Epub 2013 Sep 20
22. Deng JS, Qin P, Li XX, Du YH. Association between interleukin-1β C (3953/4)T polymorphism and chronic periodontitis: evidence from a meta-analysis. Hum Immunol. 2013;74(3):371-8 doi: 10.1016/j.humimm.2012.11.018. Epub 2012 Dec 7
23. Urbán E, Terhes G, Radnai M, Gorzó I, Nagy E. Detection of periodontopathogenic bacteria in pregnant women by traditional anaerobic culture method and by a commercial molecular genetic method. Anaerobe. 2010;16(3):283-8 doi: 10.1016/j.anaerobe.2010.02.005. Epub 2010 Feb 26
24. Ertugrul AS, Arslan U, Dursun R, Hakki SS. Periodontopathogen profile of healthy and oral lichen planus patients with gingivitis or periodontitis. Int J Oral Sci. 2013;5(2):92-7 doi: 10.1038/ijos.2013.30. Epub 2013 Jun 7
25. Reichert S, Haffner M, Keyßer G, Schäfer C, Stein JM, Schaller HG, Wienke A, Strauss H, Heide S, Schulz S. Detection of oral bacterial DNA in synovial fluid. J Clin Periodontol. 2013;40(6):591-8 doi: 10.1111/jcpe.12102. Epub 2013 Mar 28
26. Eick S, Pfister W. Comparison of microbial cultivation and a commercial PCR based method for detection of periodontopathogenic species in subgingival plaque samples. J Clin Periodontol. 2002;29(7):638-44
27. Rupf S, Kannengiesser S, Merte K, Pfister W, Sigusch B, Eschrich K. Comparison of profiles of key periodontal pathogens in periodontium and endodontium. Endodontics & Dental Traumatology 16. 2000;16(6):269-75
28. Gayathri R, Saadi AV, Bhat KM. et al. Genotype and Composite genotype effects of IL-1A+4845 and IL-1B+3954 polymorphism for chronic periodontitis in an Indian population. Indian J Dent Res. 2011;22:612
29. Cardon LR, Bell JI. Association study designs for complex diseases. Nat Rev Genet. 2001;2:91-99
30. Scapoli C, Trombelli L, Mamolini E, Collins A. Linkage disequilibrium analysis of case-control data: an application to generalized aggressive periodontitis. Genes Immun. 2005;6(1):44-52
31. Cohen P, Cohen J. The clinician's illusion. Arch Gen Psychiatry. 1984;41:1178-82
32. Armitage GC, Wu Y, Wang HY, Sorrell J, di Giovine FS, Duff GW. Low prevalence of a periodontitis associated interleukin-1 composite genotype in individuals of Chinese heritage. J Periodontol. 2000;71:164-71
33. Risch NJ. Searching for genetic determinants in the new millennium. Nature. 2000;405:847-56
34. Evidence-based dentistry. what's new? Ballini A, Capodiferro S, Toia M, Cantore S, Favia G, De Frenza G, Grassi FR. Int J Med Sci. 2007;4(3):174-8
35. Mitsiadis TA, Woloszyk A, Jiménez-Rojo L.Nanodentistry. combining nanostructured materials and stem cells for dental tissue regeneration. Nanomedicine (Lond). 2012Nov;7(11):1743-53 doi: 10.2217/nnm.12.146. Review
36. Galler KM, D'Souza RN. Tissue engineering approaches for regenerative dentistry. Regen Med. 2011;6(1):111-24 doi: 10.2217/rme.10.86
37. Mori G, Brunetti G, Ballini A, Di Benedetto A, Tarantino U, Colucci S, Grano M. Biological characteristics of dental stem cells for tissue engineering. Key Engineering Materials. 2013;541:51-59
38. Grassi FR, Ciccolella F, D'Apolito G, Papa F, Iuso A, Salzo AE, Trentadue R, Nardi GM, Scivetti M, De Matteo M, Silvestris F, Ballini A, Inchingolo F, Dipalma G, Scacco S, Tetè S. Effect of low-level laser irradiation on osteoblast proliferation and bone formation. J Biol Regul Homeost Agents. 2011;25(4):603-14
39. Ballini A, Cantore S, Capodiferro S, Grassi FR. Esterified hyaluronic acid and autologous bone in the surgical correction of the infra-bone defects. Int J Med Sci. 2009;6(2):65-71
40. Boccaccio A, Ballini A, Pappalettere C, Tullo D, Cantore S, Desiate A. Finite element method (FEM), mechanobiology and biomimetic scaffolds in bone tissue engineering. Int J Biol Sci. 2011;7(1):112-32
Author contact
![]() Corresponding author: e-mail andrea.ballinicom.
Corresponding author: e-mail andrea.ballinicom.

 Global reach, higher impact
Global reach, higher impact