Impact Factor
ISSN: 1449-1907
Int J Med Sci 2014; 11(6):620-625. doi:10.7150/ijms.8251 This issue Cite
Research Paper
PIK3CA Gene Amplification and PI3K p110α Protein Expression in Breast Carcinoma
1. Biomolecular Research Group, Biochemistry Program, Institute of Biological Sciences, Faculty of Science, University of Malaya, 50603 Kuala Lumpur, Malaysia
2. Institute of Bioscience, University Putra Malaysia, 43300 Serdang, Selangor, Malaysia
Received 2013-12-1; Accepted 2014-3-12; Published 2014-4-22
Abstract
A family of PI3Ks is the lipid kinases, which enhance intracellular pools of phosphatidyl inositol 3,4,5-tri-phosphate (PIP3) through phosphorylating its precursor. Amplifications and deletions of genes, as well as somatic missense of the PIK3CA gene have been described in many human cancer varieties, including of the brain, colon, liver, lung and stomach.
Immunohistochemistry and Real-time quantitative PCR tests were used to determine the PIK3CA gene amplification (gene copy number) and to detect protein expression, respectively. The results obtained were analysed and the ratio of PIK3CA to β-actin gene copy number was calculated. Positive gene amplification of PIK3CA was appointed as a copy number of ≥4. Also, PI3K p110α protein expression was scored from 0 to 3+ and the scores of 2+ and 3+ were considered as positive for PI3K p110α protein expression.
We studied 50 breast carcinoma samples for PI3K p110α protein expression and PIK3CA gene copy numbers. In general, 36 out of 50 (72%) breast carcinoma samples showed a significant increase in PIK3CA gene amplification. 12 out of 50 (24%) showed positive staining, and 38 out of 50 (76%) showed negative staining for PI3K p110α expression.
We have identified no significant relationship between PIK3CA amplification, race (p= 0.630) and histological type (p=0. 731) in breast carcinoma, but correlation of PIK3CA amplification and age showed a significant relationship (p=0. 003) between them.
No significant relationship has been identified in correlation of PI3K p110α protein expression compared to age (p=0. 284), race (p=0. 546) and histological type (p=0. 285).
Amplification of PIK3CA was frequent in breast carcinoma and occurs in stages of breast carcinoma. Our result shows that there is a relationship between gene amplification and age in breast carcinoma. We suggest that PIK3CA is significant in breast tumorigenesis serve as a prevalent mechanism contributes to the oncogenic activation pathway of PIK3CA in breast cancer.
Keywords: Breast cancer, PI3 kinase, Real-time PCR, Expression
Introduction
Phopsphotidylinositol 3-kinase (PI3ks) is a grouping of lipid kinases to control signalling pathways occupied in cell proliferation, adhesion, survival, and motility. Phosphatidylinositol 3-kinase (PI3K), is a main downstream signalling component of tyrosine kinases (RTKs) as a growth factor receptor. Tainted mutation and expression of PI3-kinase/AKT pathway components have been occupied in diverse fellow malignancies [1,2]. Previously published article determined PTEN and epidermal growth factor receptor (HER3) as surrogate markers in patients, which have HER2-overexpressing MBC, and it has been suggested that these markers may affect in the PI3K pathway [3]. The previous published articles establish that the PIK3CA is furthermost oncogene mutated in breast carcinoma and provision an impress for PIK3CA in carcinogenesis [4].
So far, mutations of PIK3CA stated in breast cancers, which are gathered in two main hotspot areas in exons 9 and 20, consistent to tyrosine kinase and helical domains [4,5]. In more than 80% of the cases, mutations are gathered in the helical domain and kinase domain which encoded exon 9 and exon 20, respectively. To continue, several hotspot mutations of mentioned exons specifically H1047R, E545K and E542K were demonstrated to operate the PI3K/AKT pathway over the AKT phosphorylation [6-9]. However, this study has focused mainly on the amplification of PIK3CA in breast carcinomas. We defined the occurrence ratio of PIK3CA gene copy numbers in breast carcinoma in order to make a comparison with recent published data. Lately, it has been publicized that PI3K is linked to the cellular proliferation and transformation in some cancer cell lines, which contain breast carcinoma, myelogeneous leukemia and AIDS-related Kaposi Sarcoma, prostate carcinoma and hepatocellular carcinoma [10-14]. More recently, superior PI3K protein expression and activity have been identified in glioblastomas, colorectal and lung cancer [15,16]. In consequence, these findings recommended the significance of the PI3K in the development of malignant neoplasia. For that reason, we studied the expression layout of PI3K protein expression in 50 breast carcinoma.
Material and method
Tumor samples and cell lines
Formalin-fixed, paraffin-implanted tissue samples from 50 patients have been used for this study. The tumors selected for this study were 50 breast carcinoma. All tumours histological classification was established in the World Health Organization (WHO) system.
DNA isolation
Firstly, Paraffin-embedded tissue sections were treated at room temperature for 5 minutes with xylene, pursued by digestion with Proteinase K at 56ºC for 18 hours. Genomic DNA was consequently extracted employing QIAamp® DNA Mini Kit (Qiagen, Germany). The events were achieved matching to the directions of the manufacturer's.
Polymerase Chain Reaction (PCR)
Genomic DNA was amplified by real-time PCR using the specific PIK3CA gene amplifying primers. We have used β-actin as a reference gene for control. Primers and Taqman probes were as follows, PIK3CA, 5′-AAATGAAAGCTCACTCTGGAATTCC-3' (forward), and 5′-TGTGCAATTCCTATGCAATCG-3′ (reverse), and 5′-6-carboxyfluorescein-CACTGCACTGTTAATAACTCTCAGCAGGCAAA tetramethylrhodamine-3′ (Taqman probe) [17]. Also for β-actin, 5'-TCACCCACACTGTGCCCCATCTACGA-3′ (forward), 5′-TCGGTGAGGATCTTCATGAGGTA-3′ (reverse), and 5′-6-carboxyfluorescein-ATGCCCTCCCCCATGCCATCC-tetramethylrhodamine-3′ (Taqman probe) [17]. Entire primers as well as Taqman probes were procured from Operon Biotechnologies (Aitbiotech, Singapore). Final volume of 25µl was used for PCR reaction as referred in Table 1. The PCR cycling was designed by the following steps, denaturation at 95º C for 2 minutes, 50 cycles of 95º of centigrade for thirty seconds and 60º of centigrade for forty five seconds. The comparative PIK3CA copy number of the sample was calculated by considering 2.0 as the normal diploid DNA copy number. The cutoff value of PIK3CA copy number gain was set at more or equal to 4.0 copies [18,19].
Protocol and amount of material used in real-time PCR
| X1 | X2 | |
|---|---|---|
| Taq buffer with KCL | 2.5μl | 5μl |
| MgCl2 | 3μl | 6μl |
| dNTPs | 0.5μl | 1μl |
| Forward primer | 0.75μl | 1.5μl |
| Reverse primer | 0.75μl | 1.5μl |
| Taqman probe | 0.6μl | 1.2μl |
| Taq DNA polymerase | 1.25μl | 2.5μl |
| DNA | 2μl | 2μl |
| Double-distilled water | 13.6μl | 27.2μl |
| Total volume | 25μl | 50μl |
To continue, for immunohictochemical analysis the slides were treated with 3-aminopropyltrimethoxysilane (APES). Paraffin-embedded tissue sections of thickness 4.0µm were cut using a Leica rotary microtome RM2135 (Leica Microsystems, Germany) and mounted on slides. The slides were stored at 4 ºC after drying. Briefly, the slides with paraffin sections were placed in 60ºC oven for 30 minutes. Then the tissue sections were deparaffinized and hydrated by crossing over xylene and a stratified series of ethanol. Antigen recovery as presented for 25 minutes at 100ºC in Tris-EDTA (pH9) buffer solution, into a microwave oven. After heat-induced epitope retrieval, the slides with tissue sections were allowed for 30 minutes to cool at room temperature. Then, tissue sections were washed in Tris-buffered saline with 0.1% Tween-20 (TBS-T), pH7.6. Then, incubated sections with 3% H2O2 solution for 10 minutes and after those sections were incubated in 3% BSA. After that primary antibody against PI3K (p110α 1:50) was enhanced and then incubated for 2 hours. Then, primary antibody enhancer (QIAgene) was enhanced and incubated about 20 minutes at room temperature. Then, tissues were embrocated with TBS-T and incubated in 3, 3′-diaminobenzidine (DAB) substrate chromogen system (UltraVision Plus Detection System; Thermo Fisher Scientific, Fermont, CA) for 5-20 minutes until the brown color end-products were observed and sections were washed in 2x dH2O. The tissue sections were finally counterstained with hematoxylin for 5 seconds and washed in running tap water for 2 minutes. Finally the tissues were dehydrated. Rabbit monoclonal anti-PI3K p110α antibody (clone C13F8, Cell signalling technology, USA) was used at a dilution of 1:50. Antigen detection was performed at 100ºC for 25 minutes at pH 6.0, 10mM sodium citrate buffer, in a microwave oven. Finally, tissue sections were incubated with anti-PI3K p110α antibody for one hour at room temperature.
The relationship between PIK3CA gene copy numbers and PI3K p110α protein expression and clinico-pathological records were evaluated by Fisher's exact test. Kruskal-Wallis and Mann-Whitney are the two statistical tests which were used for the correlation analysis and p˂ 0.05 was presumed as statistically significant. SPSS 20.0 parameters (SPSS Inc., USA) were used for statistical analysis.
Result
PIK3CA gene copy numbers and PI3K p110α protein expression in breast carcinoma
PIK3CA gene amplification has been studied in different type of human carcinomas, for example; uterine cancer [20], cell lung cancer [21], nasopharyngeal carcinoma [22] and endometrial cancer [23]. One of our aims was to determine the gene copy number rate of PIK3CA in breast carcinoma. Hence, genomic DNA from 50 breast carcinoma and 10 normal blood samples was used to determine the frequency of an increased copy number of PIK3CA gene. Our results suggest that, 36 out of 50 (72%) samples shows an increase in PIK3CA gene amplification in breast carcinoma and PIK3CA gene copy numbers were lower than 4, in 14 out of 50 (28%) samples. Results obtained from normal human blood samples showed that there were no significant positive gene copy numbers identified in normal blood DNA samples and PIK3CA gene copy numbers were lower than 4 in all samples.
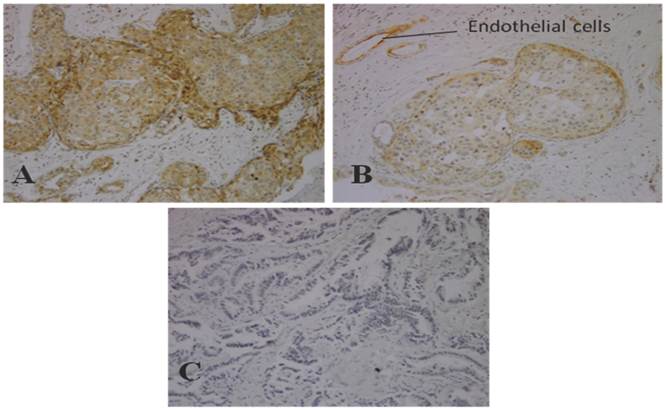
We estimated expression of PI3K p110α in breast carcinoma in 50 breast carcinoma tissues. The immunohistochemical scores of 0 and 1+ staining were considered as negative and 2+ and 3+ staining were considered positive. As results shows, 12 out of 50 (24%) of breast carcinoma samples have positive staining and 38 samples (76%) were negative. Figure 1, shows negative control, weak (1+) and moderate (2+) staining of PI3K p110α protein in breast carcinoma tissue.
Immunostaining of PI3K p110α in breast carcinoma (Original Magnification, 100x) A) Moderate staining (2+) in invasive breast cancer tissues B) Weak staining (1+) in invasive breast cancer tissues C) Negative control staining in invasive breast cancer tissues.
Relationship between PIK3CA gene copy number, PI3K p110α protein expression, age, race and histological types
In the present study, the relationship between PIK3CA gene amplification, age, race and histological type was analysed in breast carcinoma. Statistical analysis showed that there is no significant relationship between PIK3CA gene amplification, race (p=0.630) and histological type (p=0.731) in breast carcinoma, but there is a significant relationship between PIK3CA gene amplification and age (p=0.003) in breast carcinoma (Table 2).
Also, we have done correlation between PI3K p110α protein expression, age, race and histological type in breast carcinoma, which statistical analysis showed that there is no significant relationship between PI3K p110α expression, age (p=0.284), race (p=0.546) and histological type (p=0.285) in breast carcinoma (Table 3).
Correlation between PIK3CA gene copy numbers, age, race and histological types in breast carcinoma
| PI3K Copy No | Number | Mean Rank | P |
|---|---|---|---|
| Age* | |||
| =<48 | 28 | 20.21 | 0.003 |
| >48 | 22 | 32.23 | |
| Race* | |||
| Group1 | 8 | 20.00 | |
| Group2 | 28 | 25.79 | 0.630 |
| Group3 | 10 | 27.30 | |
| Group4 | 4 | 30.00 | |
| Histological Type** | |||
| I | 20 | 27.30 | |
| II | 20 | 23.70 | 0.731 |
| III | 10 | 25.50 |
*Mann-Whitney test / **Kruskal-Walley test
Correlation between PI3K p110α expression, age, race and histological grade in breast carcinoma
| PI3K p110α | Number | Mean Rank | P |
|---|---|---|---|
| Age* | |||
| =<48 | 28 | 23.79 | 0.284 |
| >48 | 22 | 27.68 | |
| Race* | |||
| Group1 | 8 | 23.50 | |
| Group2 | 28 | 25.43 | 0.546 |
| Group3 | 10 | 23.90 | |
| Group4 | 4 | 34.00 | |
| Histological type** | |||
| I | 20 | 25.80 | |
| II | 20 | 27.90 | 0.285 |
| III | 10 | 20.10 |
*Mann-Whitney test / **Kruskal-Walley test
Discussion
To find out gene copy numbers of the PIK3CA, we carried out real-time PCR over 50 breast tumor samples. Gene amplification is well accepted as a later occurrence in tumor progression [24,25].
Overall, we found a high frequency of PIK3CA gene copy numbers in breast carcinoma samples. Among the 50 cases, 36 (72%) showed a large amount of gene copy numbers in comparison to normal controls. Previously published data, specified PI3K pathway distorted in a large amount among breast carcinoma patients [26-28]. Though, the occurrence of gene copy number was much more than results identified in earlier studies (1-9-14%) [29,30]. PIK3CA amplification and mutation were nearly equally individual events and as they signify independent methods for enriching PI3K activity, an upper incidence of copy number may possibly clarify the inferior occurrence of PIK3CA amplification in this group compared to those in further reports. Among other types of cancers, genetic analysis of PIK3CA amplification were conducted in Japan, PIK3CA was over expressed in 73% of primary ovarian clear cell carcinomas [31]. In comparison with the previous published data, our data suggest that PIK3CA is significant in breast tumorigenesis, serve as a mutual mechanism contributes to the oncogenic activation of PIK3CA in breast cancer.
In breast carcinomas, PI3K over expression occurred in 24% of all tumor samples, thereby considered as not over expressed. More recently, studies informed that PI3K is not over expressed in melanocytic lesions [32]. Our data are consistent with the previous studies, whereas, p110β and p110α are all over expressed in untransformed cells and tissues, whereas p110δ expression is highly enriched in leukocytes. High levels of p110δ expression have been documented in some solid tumor cell lines, but the functional role is unknown [33]. PI3K pathway activation in breast carcinoma was distinguished using p-S6, p-AKT, and PTEN by evaluating of their expression through IHC. The expression of p-S6 and p-AKT was positive in 75% and 69% of breast carcinoma. There was No significant association between PI3K pathway status and breast carcinoma subtype for p-AKT [34].
Our data showed no significant association between PI 3-kinase expression and ages, races and histological types in breast carcinoma. Also, no relation between the PIK3CA gene copy number, race and histological type has been found, but there is a significant relation between PIK3CA gene copy number and age in breast carcinoma has reported, which may due post-transcriptional, translational and protein degradation regulation which shows some genes are produced but not expressed on the cell surface [35].
The PI3K pathway deviations are existing in a significant quantity of breast carcinoma patients. Accumulating evidence support that PI3K activation has been affiliated with persistence to ErbB2-targeted therapy, moreover standard cytotoxic therapy in breast carcinoma, and the addition of novel PI3K pathway inhibitors (e.g., PX-866, BEZ235, and XL147) which are expected to have superior therapeutic ability to treat patients resistant to standard therapies [36,37]. Although several PI3K activation biomarkers have been proposed such as p-Akt and PIP3, these biomarkers are either not specific or difficult to measure in clinical practice [38]. There was no significant correlation observed amongst PIK3CA mutation and age, differentiation, histological type and lymph node metastasis in a research has done in China [39]. Our research outcomes are not similar to the prior study, Saal et al. (2005); found positive significance between PIK3CA mutations and expression in breast cancer. We believe, variations in samples will be the reason of differences between our and the other studies, as their experiment was carried out on PIK3CA exons such as 1, 2, 4, 5, 7, 9, 12, 13, 18, 20, which were included intron-exon boundaries and their method were based on PCR as well as gene sequencing of exons so it can be the cause of inconsistency with our research. Our study also did not have any focus on mutations and we used primers for amplification of PIK3CA on exon 21. In contrast, another study done in exons 9 and 20 reported no significant relationship between PIK3CA mutations and some clinicopathologic characteristics, such as age, lymph node metastases, tumor size, ER and PR status, p53 expression and mutation in breast cancer [40].
Treatment and Survival
Based on survival statistics during of 5-year, which has been issued between 1975 to 2008 in the SEER Cancer Statistics Review; breast cancer therapy includes breast-conserving surgery (BCS) or mastectomy. Mastectomy is similar as long-term survival, when BCS is appropriately used for provincial or restricted cancers [41].
Limited of diagnosis are on age in women (20-45%), who decided to have breast reconstruction assume mastectomy elect, with each tissue flap or an implant or a mixture of the both [42-46].
Amongst women make a diagnosis in early stage (I or II) of breast carcinoma, 57% accept breast-conversing surgery, 36% assume a mastectomy , around 6% endure no surgical usage, and approximately 1% take no conduct. In contrast, across patients having late stage of breast cancer (III or IV), 13% accept breast-conserving surgery, 60% undertake mastectomy treatment, 18% assume no surgical demeanor, and 7% receive no treatment at all.
The totall comparative survival percentage in 5-year for patients (female) with breast cancer has enhanced from 75.1% to 90.0% between 1975 and 1977 to 2001 over 2007. The growth is mostly due to developments in therapy (ie, hormone therapy and chemotherapy), also to previously identified results from the extensive usage of mammography [47]. Moreover, to stage, epidermal growth factor receptor 2 (HER2) status in human, aspects which affect survival comprise hormone receptor status, and tumor grade.
Acknowledgements
Hereby, the authors would like to appreciate the University of Malaya for providing the research Grant (RP001-2012C).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Osaki M, Oshimura M, Ito H. PI3K-Akt pathway: its functions and alterations in human cancer. Apoptosis. 2004;9(6):667-76
2. Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov. 2005;4(12):988-1004
3. Park YH, Jung HA, Choi MK, Chang W, Choi YL, Do I-g, Ahn JS, Im YH. Role of HER3 expression and PTEN loss in patients with HER2-overexpressing metastatic breast cancer (MBC) who received taxane plus trastuzumab treatment. British Journal of Cancer. 2014;110:384-391
4. Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S, Konishi H, Karakas B, Blair BG, Lin C, Peters BA, Velasco VE, Park BM. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biology & Therapy. 2004;3(8):772-775
5. Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, Yu JS, Malmstorm PO, Mansukhi M, Enoksson J, Hibshoosh H, Borg A, Parsons R. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;19(3):183-232
6. Ikenoue T, Kanai F, Hikiba Y. et al. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res. 2005;65:4562-7
7. Isakoff SJ, Engelman JA, Irie HY. et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65:10992-1000
8. Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci USA. 2005;102:802-7
9. Samuels Y, Diaz LA Jr, Schmidt-Kittler O. et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561-73
10. Soldi R, Graziani A, Benelli R, Chigo D, Bosia A, Bussolino F. Oncostatin M activates phosphatidylinositol-3-kinase in Kaposi's sarcoma cells. Oncogene. 1994;9(8):2253-60
11. Skorski T, Kanakaraj P, Nieborowska-Skorska M, Ratajczak MZ, Wen SC, Zon G, Gewirtz AM, Perussia B, Calabretta B. Phosphatidylinositol-3 kinase activity is regulated by BCR/ABL and is required for the growth of Philadelphia chromosome-positive cells. Blood. 1995;86(2):726-36
12. Dufourny B, Alblas J, van Teeffelen HA, van Schaik FM, van der Brug B, Steenbergh PH, Sussenbach JS. Mitogenic signaling of insulin-like growth factor I in MCF-7 human breast cancer cells requires phosphatidylinositol 3-kinase and is independent of mitogen-activated protein kinase. J Biol Chem. 1997;272(49):31163-71
13. Thomas JE, Venugopalan M, Galvin R, Wang Y, Bokoch GM, Vlahos CJ. Inhibition of MG-63 cell proliferation and PDGF-stimulated cellular processes by inhibitors of phosphatidylinositol 3-kinase. J Cell Biochem. 1997;64(2):182-95
14. Grasso AW, Wen D, Miller CM, Rhim JS, Pretlow TG, Kung HJ. ErbB kinases and NDF signaling in human prostate cancer cells. Oncogene. 1997;15(22):2705-16
15. Phillips WA, St Clair F, Munday MD, Thomas RJ, Mitchell CA. increased levels of phosphatidylinositol 3-kinase activity in colorectal tumors. Cancer. 1998;83(1):41-7
16. Lin X, Bohle AS, Dohrmann P, Leuschner I, Schulz A, Kremer B, Fandrich F. Overexpression of phosphatidylinositol 3- kinase in human lung cancer. Langenbecks Arch Surg. 2001;386(4):293-301
17. Hou P, Liu D, Shan Y, Hu S, Studeman K, Condouris S, Wang Y, Trink A, El-Naggar AK, Tallini G, Vasko V, Xing M. Genetic alterations and their relationship in the phosphatidylinositol 3-kinase/Akt pathway in thyroid cancer. Clin Cancer Res. 2007;13(4):1161-70
18. Livak J, Schmittgen T. Analysis of Relative Gene Expression Data Using RealTime Quantitative PCR and the 2*2-ΔΔ T method. METHODS. 2001;25:402-408
19. Kita D, Yonelawa Y, Weller M, Ohgaki H. PIK3CA alterations in primary (de novo) and secondary glioblastomas. Acta Neuropathol. 2007;113(3):295-302
20. Konopka B, Janiec-Jankowska A, Kwiatkowska E, Najmoła U, Bidziński M, Olszewski W, Goluda C. PIK3CA mutations and amplification in endometrioid endometrial carcinomas: relation to other genetic defects and clinicopathologic status of the tumors. Human Pathology. 2011;42:1710-1719
21. Kawano O, Yano M, Sasaki H, Yukiue H, Okuda K, Fujii Y. A case of non-small cell lung cancer with EGFR mutation responding to S-1 after a therapy with gefitinib. Gan To Kagaku Ryoho. 2009;36(4):675-7
22. Fendri A, Khabir A, Mnejja W, Sellami-Boudawara T, Daoud J, Frikha M, Ghorbel A, Gargouri A, Mokdad-Gargouri R. PIK3CA amplification is predictive of poor prognosis in Tunisian patients with nasopharyngeal carcinoma. Cancer Sci. 2009;100(11):2034-9
23. Miyake T, Yoshino K, Enomoto T. PIK3CA gene mutations and amplifications in uterine cancers, identified by methods that avoid confounding by PIK3CA pseudogene sequences. Cancer Lett. 2008;261:120-6
24. Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643-649
25. Gray JW, Collins C. Genome changes and gene expression in human solid tumors. Carcinogenesis. 2000;21:443-452
26. Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo WL, Davies M, Carey M, Hu Z, Guan Y, Sahin A, Symmans WF, Pusztai L. et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68(15):6084-91
27. Bose S, Chandran S, Mirocha JM, Bose N. The Akt pathway in human breast cancer: a tissue-array-based analysis. Mod Pathol. 2006;19:238-45
28. Tokunaga E, Kimura Y, Mashino K, Oki E, Kataoka A, Ohno S, Morita M, Kakeji Y, Baba H, Maehara Y. Activation of PI3K/ Akt signaling and hormone resistance in breast cancer. Breast Cancer. 2006;13:137-44
29. Campbell IG, Russell SE, Choong DY, Montgomery KG, Ciavarella ML, Hooi CS, Cristiano BE, Pearson RB, Phillips WA. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64(21):7678-81
30. Wu G, Xing M, Mambo E, Huang X, Liu J, Guo Z, Chatterjee A, Goldenberg D, Gollin SM, Sukumar S, Trink B, Sidransky D. Somatic mutation and gain of copy number of PIK3CA in human breast cancer. Breast Cancer Res. 2005;7(5):R609-16
31. Abe A, Minaguchi T, Ochi H, Onuki M, Okada S, Matsumoto K, Satoh T, Oki A, Yoshikawa H. PIK3CA overexpression is a possible prognostic factor for favorable survival in ovarian clear cell carcinoma. Hum Pathol. 2013;44(2):199-207
32. Singh RS, Diwan AH, Zhang PS, Prieto VG. Phosphoinositide 3-kinase is not overexpressed in melanocytic lesions. J Cutan Pathol. 2007;34(3):220-5
33. Tzenaki N, Andreou M, Stratigi K, Vergetaki A, Makrigiannakis A, Vanhaesebroeck B, Papakonstanti EA. High levels of p110δ PI3K expression in solid tumor cells suppress PTEN activity, generating cellular sensitivity to p110δ inhibitors through PTEN activation. FASEB J. 2012;26(6):2498-508
34. Barbara A, Allison MD, Emily B, Joseph G, Erika H, Kimberly LB, Chad L, Karen F, Aleix P, J C Harrell, Matthew GE, Lisa AC, C R Miller, Carey KA. Phosphatidylinositol 3-kinase pathway activation in breast cancer brain metastases. licensee BioMed Central Ltd. 2001;13:R125
35. Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13(4):227-32
36. Bedard PL, Cardoso F, Piccart-Gebhart MJ. Stemming resistance to HER-2 targeted therapy. J Mammary Gland Biol Neoplasia. 2009;14:55-66
37. Kolasa IK, Rembiszewska A, Felisiak A, Ziolkowska-Seta I, Murawska M, Moes J, Timorek A, Dansonka-Mieszkowska A, Kupryjanczyk J. PIK3CA amplification associates with resistance to chemotherapy in ovarian cancer patients. Cancer Biol Ther. 2009;8(1):21-6
38. Coughlin CM, Johnston DS, Strahs A, Burczynski ME, Bacus S, Hill J. et al. Approaches and limitations of phosphatidylinositol-3-kinase pathway activation status as a predictive biomarker in the clinical development of targeted therapy. Breast Cancer Res Treat. 2010;124:1-11
39. Li SY, Wang W, Li JM, Wang Z, Wen RY, Chen J, Miao XT. PIK3CA mutation is an independent indicator of malignant phenotype and prognosis in breast cancer. Zhonghua Zhong Liu Za Zhi. 2011;33(8):605-8 [Article in Chinese]
40. Buttitta F, Felicion L, Barassi F, Martella C, Paolizzi D, Fresu G, Salvatore S, Cuccurullo F, Mezzeti A, Campani D, Marchetti A. PIK3CA mutation and histological type in breast carcinoma: high frequency of mutations in lobular carcinoma. J Pathol. 2006;208(3):305-5
41. Jatoi I, Proschan MA. Randomized trials of breast-conserving therapy versus mastectomy for primary breast cancer: pooled analysis of updated results. Am J Clin Oncol. 2005;28:289-294
42. Lee CN, Belkora J, Chang Y, Moy B, Partridge A, Sepucha K. Are patients making high-quality decisions about breast reconstruction after mastectomy? [outcomes article]. Plast Reconstr Surg. 2011;127:18-26
43. Alderman AK, Hawley ST, Waljee J, Morrow M, Katz SJ. Correlates of referral practices of general surgeons to plastic surgeons for mastectomy reconstruction. Cancer. 2007;109:1715-1720
44. Alderman AK, Wei Y, Birkmeyer JD. Use of breast reconstruction after mastectomy following the Women's Health and Cancer Rights Act. JAMA. 2006;295:387-388
45. Christian CK, Niland J, Edge SB. et al. A multi-institutional analysis of the socioeconomic determinants of breast reconstruction: a study of the National Comprehensive Cancer Network. Ann Surg. 2006;243:241-249
46. Tseng JF, Kronowitz SJ, Sun CC. et al. The effect of ethnicity on immediate reconstruction rates after mastectomy for breast cancer. Cancer. 2004;101:1514-1523
47. Berry DA, Cronin KA, Plevritis SK. et al. Effect of screening and adjuvant therapy on mortality from breast cancer. N Engl J Med. 2005;353:1784-1792
Author contact
![]() Corresponding author: Habsah Abdul Kadir. Biomolecular research group, Biochemistry Program, Institute of Biological Sciences, Faculty of Science Building, University of Malaya, 50603 Kuala Lumpur, Malaysia. Tel. (0060) 176788101; E-mail: habsahedu.my
Corresponding author: Habsah Abdul Kadir. Biomolecular research group, Biochemistry Program, Institute of Biological Sciences, Faculty of Science Building, University of Malaya, 50603 Kuala Lumpur, Malaysia. Tel. (0060) 176788101; E-mail: habsahedu.my

 Global reach, higher impact
Global reach, higher impact