Impact Factor
ISSN: 1449-1907
Int J Med Sci 2014; 11(5):488-493. doi:10.7150/ijms.6764 This issue Cite
Research Paper
Identifying Hedgehog Signaling Specific MicroRNAs in Glioblastomas
Department of Neurosurgery, Huashan Hospital, Fudan University, Shanghai, 200040, China.
* These authors contribute equally to this work.
Received 2013-5-26; Accepted 2014-3-11; Published 2014-3-21
Abstract
Aberrant activation of hedgehog (Hh) signaling pathway plays an important role in the development and proliferation of glioblastoma (GBM) cells. However, its mechanism remains unknown. MicroRNAs (miRNAs) are short non-coding RNA molecules which are involved in the post-transcriptional regulation of genes, and enrolled in signaling transduction network in tumors. This study was designed to investigate the role of miRNAs targeting the Hh signaling pathway in GBMs. According to the expression level of Gli1 mRNA measured by real time PCR, GBM samples were assigned to Gli1 high or low expression group. MiRNA microarray was applied to screen the dysregulated miRNA. As a result, 17 miRNAs were differentially expressed between Gli1 high expression and low expression groups (p < 0.005). Thirteen miRNAs including miR-125b-1 were downregulated, while only 4 miRNAs including miR-144 were upregulated in Gli1 high expression group. In summary, our study presents a subset of miRNAs which target the Hh signaling pathway in GBMs, and throws some light on the aberrant activation mechanism.
Keywords: MicroRNA, Glioblastoma, Hedgehog, Signaling pathway.
Introduction
Glioblastoma (GBM) is the most common primary brain tumor in adults, with an incidence rate about 0.0355‰ per year [1]. GBM is characterized by remarkable biological heterogeneity, active proliferation and invasion, and poor outcome. Despite current advances in multimodal therapies, such as surgery, radiotherapy and chemotherapy, the outcome of GBM patients remain unsatisfied [2], which may be partially due to the incomplete understanding of the multiple mechanisms of tumors.
Since the discovery of the Drosophila Hedgehog (Hh) [3] in 1980, Hh signaling pathway has been found to play multiple roles in tumorigenesis, development and homeostasis [4-7]. Hh signaling pathway is critical for embryonic patterning and development in the central nervous system (CNS) [6, 8]. Recent data have shown that Hh signaling pathway is involved in the initiation and maintenance of GBM [9-12]. Aberrant activation of the Hh signaling pathway may result in tumorigenesis of human cancer, and also play an important role in proliferation of neoplastic cells [13, 14]. While inactivation of the Hh signaling pathway leads to depletion of stem cell capacity in glioblastoma [15, 16]. Experiments have indicated that overexpression of Gli1 in the CNS of tadpole's leads to tumor formation [17]. Such aberrant activation is often resulted from the mutation of PTCH1 (as in BCCs (basal cell carcinoma), familial Gorlin's or basal cell nevus syndrome), SMO or even other components of the pathway, such as suppressor of fused (Sufu) [14, 18]). However, the PTCH1 mutation has not been reported in GBMs so far.
Although an important effect on the pathway deregulation has been demonstrated for various tumors, they do not account for all types of activation. More specifically, translational control of Hh pathway components and its deregulation in GBMs have not been systematically investigated yet.
Recently, microRNAs (miRNAs) have been implicated in tumor formation [19] as well as in the control of neural cell development [20]. MiRNAs are a class of small non-coding single-stranded RNAs that regulate a wide spectrum of gene expression by binding to the 3' untranslated regions (3'UTRs) of target mRNAs, resulting in their translational inhibition or degradation [21]. Recent studies have identified dysregulation of specific miRNAs in malignant gliomas [22]. These observations suggest the potential involvement of misregulated miRNAs in the malignant transformation. However, little is known about the miRNA-mediated specific targeting of developmental pathways (i.e. Hh) involved in tumor formation.
In this study, a high-throughput miRNA microarray screening of miRNA expression in subsets of human GBMs allowed us to identify the specific miRNAs involved in the regulation of the Hh pathway: 13 miRNAs were down-regulated, and 4 miRNAs were up-regulated.
Materials and Methods
Human samples
Twelve human primary glioblastoma specimens were collected during surgical resection in our department. Human GBMs were divided into two subsets: Gli1 high expression group that exhibited Gli1 levels significantly exceeding the average adult cerebella and Gli1 low expression group that presented levels in the normal range or below.
RNA isolation
Total RNA was isolated using TRIzol (Invitrogen) and miRNeasy mini kit (QIAGEN) according to manufacturer's instructions, which efficiently recovered all RNA species, including miRNAs. RNA quality and quantity was measured by using nanodrop spectrophotometer (ND-1000, Nanodrop Technologies) and RNA Integrity was determined by gel electrophoresis.
cDNA synthesis and real-time PCR
cDNA synthesis was performed using the Gene Amp PCR System 9700 (Applied Biosystems) according to the manufacturer's instructions. Quantitative real time PCR analysis of Gli1 and GAPDH mRNA expression was analyzed on Rotor-Gene 3000 real time PCR system (Corbett Research) according to the manufacturer's instructions. Each amplification reaction was performed in triplicate. The average of the three threshold cycles was used to calculate the amount of transcripts in the sample (SDS software, ABI). mRNA quantification was expressed, in arbitrary units, as the ratio of the sample quantity to the calibrator or to the mean values of control samples. All values were normalized to the endogenous control GAPDH.
miRNA microarray
The 6th generation of miRCURY LNA Array (v.16.0) (Exiqon) contains more than 1891 capture probes, covering all human, mouse and rat microRNAs annotated in miRBase 16.0, as well as all viral microRNAs related to these species. In addition, this array contains capture probes for 66 new miRPlus human microRNAs. After RNA isolation from the samples, the microarray labeling and array hybridization procedure were performed according to the manufacturer's instructions. Following hybridization, the slides were achieved, and finally dried by centrifugation. Then the slides were scanned using the Axon GenePix 4000B microarray scanner (Axon Instruments, Foster City, CA).
Cell cultures and viable cell number counting
A172 glioma cells were cultured in MEM medium (Gibco, CA), with 20% fetal bovine serum at 37℃ in an atmosphere of 5% CO2, and the media were replaced routinely. miRNA mimics were transfected into the A172 glioma cells using lipofectamine (Invitrogen, USA) according to the manufacturer's protocol. The relative viable cell numbers were measured by the MTT assay 24, 48, 72 and 96 hours after transfection, according to the manufacturer's protocol. And the relative viable cell numbers were measured as the ratio according to the cell numbers at the 24h time point.
Western blot assay
Forty-eight hours after transfection, cells were harvested and lysed in a solution containing Tris-HCl pH 7.6, 50mM deoxycholic acid, sodium salt 1%, NaCl 150mM, NP40 1%, EDTA 5mM, NaF 100mM and protease inhibitors. Lysates were separated on SDS-PAGE and immunoblotted using standard procedures. Anti-Gli1 (Santa Cruz Biotechnology), anti-actin (Santa Cruz Biotechnology) and HRP-conjugated secondary antisera (Santa CruzBiotechnology) were used. Bands were visualized by EZ-ECL detection reagents.
Data analysis
Scanned images were then imported into GenePix Pro 6.0 software (Axon) for grid alignment and data extraction. Replicated miRNAs were averaged and the miRNAs with intensities >= 50 in all samples were chosen for calculating normalization factor. Expressed data were normalized using the median normalization. After normalization, significant differentially expressed miRNAs were identified through Volcano Plot filtering. Hierarchical clustering was performed using MEV software (v4.6, TIGR).
Results and Discussion
It is noted that most GBMs have an activated functional Hh pathway [9, 11]. Hh signaling pathway is triggered by the binding of the Hh extracellular proteins to the 12-pass transmembrane protein Patched receptor (Ptch), which normally inhibits the activity of 7-pass transmembrane protein Smoothened (Smo). Relieving inhibitory activity upon the Smo inside the cell, in turn activates the Hh signal pathway by a complex of proteins through intricate mechanisms, leading to the action of the zinc-finger Gli transcription factors. Gli1 is the most important factor among the three Gli factors, namely Gli1, Gli2, Gli3, though it is originally identified as an amplified gene in a human glioma line [23]. Gli1 is both the downstream effector and target gene of the pathway [24-26]. Moreover, Gli1 is a sensitive read-out of the Hh pathway activity.
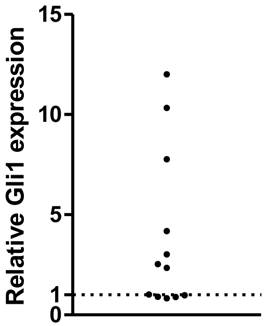
The expression of Gli1 mRNA was analyzed using RT-PCR in a series of human primary GBMs. After normalized to the GAPDH expression as endogenous control, the Gli1 mRNA expression was measured. As a result, a subset of them, 7 out of 12 samples (58%) displayed higher levels of Gli1, while the rest 5 of the 12 GBMs were considered with low Gli1 activity, which was consistent with our previous study. The 12 GBMs were then divided into Gli1 high expression group and low expression group (Figure 1).
Relative Gli1 expression in glioblastoma tissues( each plot represent one tissue ).
Previous studies have showed that Hh signaling pathway is highly related to the tumor genesis and proliferation. PTCH1 and SMO mutation were thought to be the likely mechanism in a set of tumors, including MB (medulloblastoma), BBC [14, 18], but its role remains unclear in GBMs. Recent studies have revealed that miRNAs are crucial post-transcriptional regulators of gene expression and control of cell differentiation and proliferation. More and more miRNAs were found to be related to the genesis and proliferation of GBMs [27]. However, little is known about their targeting of Hh signaling pathway in GBMs.
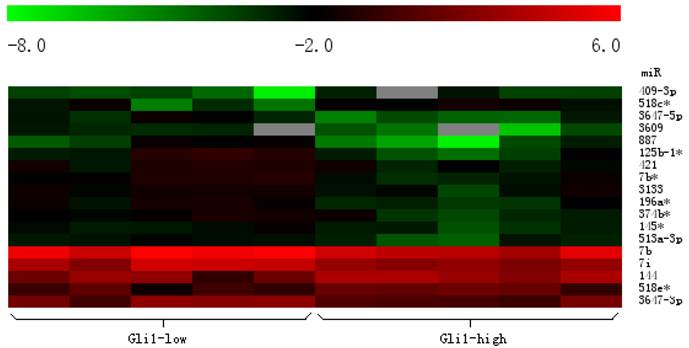
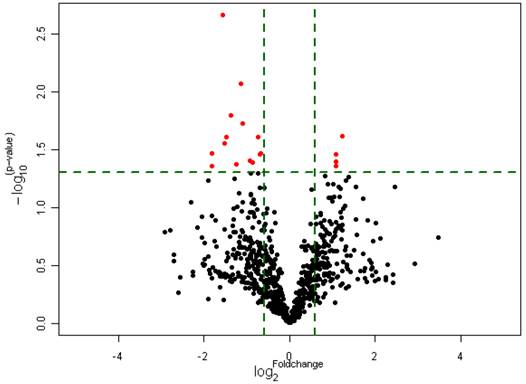
To investigate whether the expression of miRNAs varies with the functional status of Hh signaling pathway in GBMs, we carried out a microarray analysis using a microarray chip able to examine the global expression levels of 1891 probe miRNAs. Each probe has four repeat spots on the microarray. By this method, we analyzed 10 cases of primary GBM specimens, 5 cases for each group. Following assay optimization, the expression of miRNAs in GBM samples was compared between Gli1 high expression and low expression groups. Seventeen miRNAs were differentially expressed between the two groups (p < 0.005, the fold change cutoff value is 1.5). Thirteen miRNAs including miR-125b-1 were found down-regulated in GLI1 high-expression tumors, and miR-125b-1 showed the highest fold change of 1.7 to 2.5. Only 4 miRNAs including miR-144 were up-regulated, among them miR-144 is the upmost differentially expressed miRNA with 2.3 fold change. (see Figure 2, Figure 3 and Table 1)
Though the microRNA microarray has its own internal control, RT-PCR of miRNAs were carried out to test the validity of the miRNA array results. Real time RT-PCR demonstrated that miR-144 expression was up-regulated in Gli1 high GBMs, and miR-125b-1 was down-regulated. The result was consistent with that of miRNA microarray.
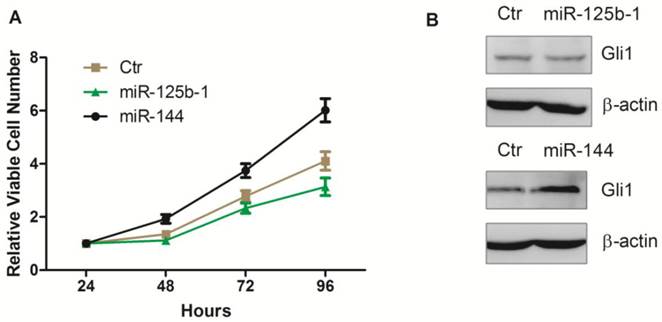
Next, we investigated whether these miRNAs mediated regulation of Hh pathway also leads to regulation in cell proliferation. We first transfected the ectopic expression of miR-125b-1 and miR-144 into A172 glioma cells. Notably, we observed that ectopic expression of miR-144 led to an increase of viable cell numbers in A172 glioma cell, while ectopic expression of miR-125b-1 showed a slight decrease effect only at the 96h time point (Figure 4A). Western blot was performed 48h after the transfection, to test the Gli1 expression. There was no significant difference between Gli1 expression before and 48h after miR-125b-1 mimic transfection, consistent with the result of viable cell numbers at the 48h time point, suggesting that the miR-125b-1 effect in A172 glioma cells might not be dominant. However, overexpression of miR-144 led to an increase of Gli1 expression (Figure 3B). As the expression of miR-125b-1 and miR-144 were regulated in human glioblastomas together with different levels of Hh signaling, we hypothesized that these miRNAs may be responsible for the the regulation of Hh activity and the consequent cell proliferation.
List of miRNAs differential expression of miRNAs in Gli1 high versus Gli1 low GBMs.
| Up regulated | miR-144, miR-409-3p, miR-518c*, miR-518e* |
|---|---|
| Down regulated | let-7b, let-7i, miR-125b-1, miR-145*, miR-196a*, miR-374b*, miR-421, miR-513a-3p, miR-887, miR-3133, miR-3609, miR-3647-5p, miR-3647-3p |
Heat map clustering of differential expression of miRNAs in Gli1 high versus Gli1 low GBMs. (Red indicates high relative expression, and green indicates low relative expression).
Differential expression of miRNAs in Gli1 high versus Gli1 low GBMs which passed Volcano Plot. (Fold change 1.5, p<0.005) (the Red spots represents the target miRNAs).
Ectopic overexpression of miR-125b-1 and miR-144 in A172 glioma cells. A. Relative viable cell numbers in A172 cells after transfection with miR-125b-1 or miR-144 or miRNA mimic negative control(ctr). B. Western blot analysis of Gli1 (upper panel) or ß-actin (bottom panel) protein levels in A172 cells 48 h after transfection with miR-125b-1 or miR-144 or compared with scrambled control (ctr).
Extensive modulation of miRNAs in GBMs was firstly studied by Ciafre in 2005, using microarray to examine the global expression levels of 245 miRNAs in GBMs. They found that miRNAs could be divided into groups, depending on their differential expression, representing some of the molecular markers of GBMs [28]. That is, a selected group of miRNAs can be exploited to disclose distinct patterns of expression likely reflecting some of the mechanisms involved in GBM tumorigenesis or more widely in the de-differentiation process often characterizing tumor growth. The miR-181 family, miR-181a, b, and c, were generally under expressed according to Ciafre study, which later were confirmed by Shi [29], and miR181a was negatively correlated with glioma grading. Furthermore, transfection of miR-181a and miR-181b inhibited the tumor proliferation, invasion, and triggered apoptosis. Similarly, most of the altered miRNAs discovered by Ciafre were confirmed by other experiments and their mechanisms were revealed. However, more than 1000 miRNAs were discovered in human cells until recently, many other miRNAs were also found to be related to some characteristics of GBMs beyond Ciafre's results, such as miR-129, miR-139, miR-124, miR-137, miR-7, etc. (summarized in Novakova's review [30]).
In most studies, GBMs were directly compared to the normal tissue or cell lines. It is easy to find the unique features of GBMs. But the disadvantage is apparent, for GBMs have multiple genetic mechanisms, and GBMs per se may be different in many specific aspects. Many signaling pathways are believed to be crucial to GBMs, including EGFR, Wnt, Hh and Notch pathways [31]. Moreover, many miRNAs were found to be targeted in the above signaling pathway. But, little was known about the relationship between miRNA and Hh pathway in GBMs, let alone the systematic investigation. So, this study was conducted to explore the specific miRNAs targeting the Hh pathway in GBMs.
Notably, this result dramatically differed from Ciafre's report. In our study, with the help of 1891 probe miRNAs, we found some miRNAs for the first time, which may be the biomarkers of a subset of GBMs. Our microarray analysis detected a significant change of 17 miRNAs including miR-144, miR-125b-1, and miR-421. The 17 miRNAs may be the specific miRNAs which target the Hh pathway in GBMs. Among these miRNAs, miR-125b has been reported as a suppressor of Hh signal pathway in medulloblastomas (MBs). The lowest expression of miR-125b was observed in Gli1 high-expression MBs, while over-expression of miR-125b targeted the activator components of the Hh signaling pathway (Smo). Besides, the authors pointed out that miR-125b, miR-326 together with miR-324-5p targeted the main component of Hh signaling pathway, suggesting that several miRNAs may be required to efficiently impair the oncogenic potential of the Hh pathway [32]. Therefore, our results, though not precise, may be a good choice for further screening of the miRNAs which activate the Hh signaling pathway in GBMs.
Although we described a subset of miRNAs which may control the Hh pathway in GBMs, we had not found the specific miRNAs associated with the tumorigenesis in GBMs. In further study, we plan to regulate the expression of miRNAs to confirm the combination of miRNAs and its targets in Hh signaling pathway components.
In conclusion, a subset of miRNAs related to the aberrant activation of Hh signaling pathway are identified in GBMs by using the microarray chip. Additional studies are needed to experimentally characterize the altered miRNAs and their target mRNA in Hh signaling pathway.
Acknowledgements
This study was funded by grants from the Natural Science Foundation of Shanghai (No. 11ZR1405100).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. Journal of Neuropathology and Experimental Neurology. 2005;64:479-89
2. Siebzehnrubl FA, Reynolds BA, Vescovi A, Steindler DA, Deleyrolle LP. The Origins of Glioma: E Pluribus Unum? Glia. 2011;59:1135-47 doi:10.1002/glia.21143
3. Nussleinvolhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795-801 doi:10.1038/287795a0
4. Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes & Development. 2001;15:3059-87 doi:10.1101/gad.938601
5. Mullor JL, Sanchez P, Altaba ARI. Pathways and consequences: Hedgehog signaling in human disease. Trends Cell Biol. 2002;12:562-9 doi:10.1016/s0962-8924(02)02405-4
6. Rubin LL, de Sauvage FJ. Targeting the Hedgehog pathway in cancer. Nat Rev Drug Discov. 2006;5:1026-33 doi:10.1038/nrd2086
7. Marini KD, Payne BJ, Watkins DN, Martelotto LG. Mechanisms of Hedgehog signalling in cancer. Growth Factors. 2011;29:221-34 doi:10.3109/08977194.2011.610756
8. Altaba ARI. Hedgehog Signaling and the Gli Code in Stem Cells, Cancer, and Metastases. Sci Signal. 2011 doi:pt910.1126/scisignal.2002540
9. Wang K, Pan L, Che XM, Cui DM, Li C. Gli1 inhibition induces cell-cycle arrest and enhanced apoptosis in brain glioma cell lines. J Neuro-Oncol. 2010;98:319-27 doi:10.1007/s11060-009-0082-3
10. Cui DM, Xu QW, Wang K, Che XM. Gli1 is a potential target for alleviating multidrug resistance of gliomas. J Neurol Sci. 2010;288:156-66 doi:10.1016/j.jns.2009.09.006
11. Braun S, Oppermann H, Mueller A, Renner C, Hovhannisyan A, Baran-Schmidt R. et al. Hedgehog signaling in glioblastoma multiforme. Cancer Biol Ther. 2012;13:487-95 doi:10.4161/cbt.19591
12. Takezaki T, Hide T, Takanaga H, Nakamura H, Kuratsu J, Kondo T. Essential role of the Hedgehog signaling pathway in human glioma-initiating cells. Cancer Sci. 2011;102:1306-12 doi:10.1111/j.1349-7006.2011.01943.x
13. Villavicencio EH, Walterhouse DO, Iannaccone PM. The Sonic hedgehog-Patched-Gli pathway in human development and disease. American Journal of Human Genetics. 2000;67:1047-54 doi:10.1016/s0002-9297(07)62934-6
14. Cohen MM Jr. Hedgehog Signaling Update. American Journal of Medical Genetics Part A. 2010;152A:1875-914 doi:10.1002/ajmg.a.32909
15. Bar EE, Chaudhry A, Lin A, Fan X, Schreck K, Matsui W. et al. Cyclopamine-mediated hedgehog pathway inhibition depletes stem-like cancer cells in glioblastoma. Stem Cells. 2007;25:2524-33 doi:10.1634/stemcells.2007-0166
16. Clement V, Sanchez P, de Tribolet N, Radovanovic I, Altaba ARI. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol. 2007;17:165-72 doi:10.1016/j.cub.2006.11.033
17. Dahmane N, Sanchez P, Gitton Y, Palma V, Sun T, Beyna M. et al. The Sonic Hedgehog-Gli pathway regulates dorsal brain growth and tumorigenesis. Development. 2001;128:5201-12
18. Ruiz i Altaba A, Palma V, Dahmane N. Hedgehog-Gli signalling and the growth of the brain. Nature Reviews Neuroscience. 2002;3:24-33 doi:10.1038/nrn704
19. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857-66 doi:10.1038/nrc1997
20. Kosik KS. The neuronal microRNA system. Nature Reviews Neuroscience. 2006;7:911-20 doi:10.1038/nrn2037
21. Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nature Reviews Molecular Cell Biology. 2008;9:219-30 doi:10.1038/nrm2347
22. Katakowski M, Buller B, Wang X, Rogers T, Chopp M. Functional MicroRNA Is Transferred between Glioma Cells. Cancer Res. 2010;70:8259-63 doi:10.1158/0008-5472.can-10-0604
23. Kinzler KW, Bigner SH, Bigner DD, Trent JM, Law ML, Obrien SJ. et al. Identification of an amplified, highly expressed gene in a human glioma. Science. 1987;236:70-3 doi:10.1126/science.3563490
24. Altaba ARI. The works of GLI and the power of Hedgehog. Nat Cell Biol. 1999;1:E147-E8
25. Ho KS, Scott MP. Sonic hedgehog in the nervous system: functions, modifications and mechanisms. Current Opinion in Neurobiology. 2002;12:57-63 doi:10.1016/s0959-4388(02)00290-8
26. Nybakken K, Perrimon N. Hedgehog signal transduction: recent findings. Current Opinion in Genetics & Development. 2002;12:503-11 doi:10.1016/s0959-437x(02)00333-7
27. Turner JD, Williamson R, Almefty KK, Nakaji P, Porter R, Tse V. et al. The many roles of microRNAs in brain tumor biology. Neurosurg Focus. 2010 doi:E310.3171/2009.10.focus09207
28. Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G. et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochemical and Biophysical Research Communications. 2005;334:1351-8 doi:10.1016/j.bbrc.2005.07.030
29. Shi L, Cheng ZH, Zhang JX, Li R, Zhao P, Fu Z. et al. hsa-mir-181a and hsa-mir-181b function as tumor suppressors in human glioma cells. Brain Res. 2008;1236:185-93 doi:10.1016/j.brainres.2008.07.085
30. Novakova J, Slaby O, Vyzula R, Michalek J. MicroRNA involvement in glioblastoma pathogenesis. Biochemical and Biophysical Research Communications. 2009;386:1-5 doi:10.1016/j.bbrc.2009.06.034
31. Dietrich J, Diamond EL, Kesari S. Glioma stem cell signaling: therapeutic opportunities and challenges. Expert Rev Anticancer Ther. 2010;10:709-22 doi:10.1586/era.09.190
32. Ferretti E, De Smaele E, Miele E, Laneve P, Po A, Pelloni M. et al. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. Embo J. 2008;27:2616-27 doi:10.1038/emboj.2008.172
Author contact
![]() Corresponding author: Department of Neurosurgery, Huashan Hospital, Fudan University, Shanghai, 200040, China. Fax: +86-21-52887210. Email: 0456144edu.cn.
Corresponding author: Department of Neurosurgery, Huashan Hospital, Fudan University, Shanghai, 200040, China. Fax: +86-21-52887210. Email: 0456144edu.cn.

 Global reach, higher impact
Global reach, higher impact