Impact Factor
ISSN: 1449-1907
Int J Med Sci 2013; 10(3):235-242. doi:10.7150/ijms.5524 This issue Cite
Research Paper
An Association Study on Genetic Polymorphisms of Rab37 Gene with the Risk of Esophageal Squamous Cell Carcinoma in a Chinese Han Population
1. Department of Medical Genetics, Third Military Medical University, Chongqing 400038, People's Republic of China;
2. Department of Thoracic and Cardiac Surgery, Southwest Hospital, Third Military Medical University, Chongqing 400038, People's Republic of China.
Received 2012-11-11; Accepted 2013-1-13; Published 2013-1-18
Abstract
Background: Rab37 encodes a Rab GTPase which regulates the vesicular transport of exocytosis. But the different findings in two types of cancers made its roles in oncology more confused. In this study, a clinical research on genetic polymorphisms was performed to evaluate the association between Rab37 and esophageal squamous cell carcinoma (ESCC).
Methods: The mRNA expression was tested by reverse transcription-polymerase chain reaction (RT-PCR) in four ESCC cell lines. A case-control study including 212 ESCC patients and 213 cancer-free controls was genotyped by PCR-restriction fragment length polymorphism. The Hardy-Weinberg equilibrium test, association analysis and haplotype analysis were performed with SPSS and SHEsis software respectively.
Results: Rab37 mRNA could be specifically detected in two ESCC cell lines, EC109 and EC9706, but not in KYS150 and KYS450. The allele, genotype and haplotype frequencies of rs9904078G>A, rs2034310T>C and rs5018106T>C, located in Rab37, did not significantly differ between the patients and the controls. No association between the polymorphisms and the TNM stages of patients was found.
Conclusions: Rab37 mRNA was specifically expressed in some ESCC cell lines but its genetic polymorphisms were not associated with ESCC.
Keywords: Rab37, Esophageal squamous cell carcinoma, Polymorphisms, Case-control study.
Introduction
Rab37 gene encodes a Rab GTPase protein, belonging to the functional group III of the Rab family [1]. It was first reported that Rab37 played an important role in the exocytosis of the dense core granules from the mast cells [2]. It was also associated with the secretory granules of insulin in rat insulin-secreting cell line, the dense-core vesicles in neuroendocrine PC12 cells and the TNF-α secretion from activated macrophages [3-5]. Because of its principal roles in vesicular transports, it was hypothesized whether Rab37 would be involved in cell-cell and cell-extracellular matrix interactions and then probably in the tumorigenesis, invasion and metastasis. Although it was firmly believed to be involved in the intracellular traffics from trans-Golgi to plasma membrane, the functions of Rab37 in oncology were not well unknown. And two different findings even made its roles more confused. Rab37 and its interacting protein, TMEM22 (transmembrane protein 22), were up-regulated in renal cell carcinoma (RCC) clinical samples and cell lines. Knockdown of its expression caused significant reduction of cancer cell growth, suggesting that Rab37 was important for the survival of RCC cells [6]. But the lower Rab37 mRNA expression was found in the most non-small cell lung cancer (NSCLC) patients and its knockdown led to a significant increase in cell migration. Then, Rab37 was regarded as a metastasis-related tumor suppressor gene (TSG) in lung cancer [7]. Except the two reports, there has been none information on the relationship between Rab37 and the other cancer till now. Therefore, it's hard to evaluate or understand the biological functions of Rab37 in cancers.
Esophageal cancer is one of the most common causes of cancer-related mortality worldwide and its incidence rate has been significantly increasing recent years [8]. It has two different forms: esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EA) [9]. In China, ESCC is one of the most vital and extremely fatal diseases, compared with European and American nations where EA has a higher incidence [10]. Despite attempts to improve outcomes with multi-therapy means, prognosis remains very poor with the 2-year and 5-year overall survival of 35-42% and 15-24%, respectively [11]. Thus, the investigations on identification of the risk factors involved in esophageal cancer become more meaningful. Smoking was confirmed as a most important environmental risk factor for ESCC [12], whereas more and more genetic risk factors were gradually disclosed to be involved in esophageal cancer aetiology. To find these genetic factors, the association analysis was proven to be a powerful method. As previous work shown, we had demonstrated that survivin, decoy receptor 3, microRNA-196a and microRNA-146a were associated with the risk of ESCC in Chinese Han population [13-16].
In this report, we collected an ESCC case-control sample pool, tested the expression of Rab37 in different cell lines, and then performed the genotyping analysis for its Tag single nuclear polymorphisms (SNPs) to evaluate whether it would be associated with the susceptibility and the clinic stage of ESCC.
Materials and methods
Cell lines and cell cultures
KYS150, KYS450, EC109, EC9706 and Huh7 were from Tumor Center of Chinese Academy of Medical Science. EA.hy926, HepG2, MCF7 and SW1990 were from American Type Culture Collection (ATCC). All cell lines were cultured in Dulbecco's modified Eagle's minimum essential medium (DMEM, Gibco) with 10% fetal bovine serum (FBS, Gibco) at 37°C in 5% CO2.
Study populations
This case-control study had been approved by the Ethics Committee of Third Military Medical University. All subjects were genetically unrelated ethnic Han Chinese and from Chongqing City of southwest China. The sample pool consisted of 212 ESCC patients and 213 cancer-free controls, which were recruited between 2005 and 2011 at the Southwest Hospital in Chongqing, China. The ESCC patients were pathologically diagnosed and confirmed at the hospital. The exclusion criteria included previous cancer, metastasized cancer and family history of cancer. The TNM stage subdivision of esophageal cancer patients conformed to the American Joint Committee on Cancer TNM Classification of Carcinoma. The cancer-free controls, visiting the same hospital for physical examination, had no personal or family history of cancer and other genetic disease and frequency matched to the cases on gender, age and residential area.
Nucleic acid isolation
Genomic DNA was isolated from fresh blood by Wizard Genomic DNA PuriWcation Kit (Promega) according to the manufacturer's protocol. Total RNA was isolated from fresh tissue and cell lines by RNAiso Plus (TaKaRa) following the manufacturer's instructions. The quantity and quality of nucleic acid were determined by NanoDrop 1000 (Theremo).
Reverse transcription-polymerase chain reaction (RT-PCR)
Reverse transcription and PCR were performed using the SYBR PrimeScript RT Kit as described by the manufacturer (TaKaRa). GAPDH was chosen as the reference gene and its primer pairs were a sense primer 5'AACGGATTTGGTCGTATTGGG3' and an antisense primer 5'TCGCTCCTGGAAGATGGTGAT3', with PCR product of 216 bp. The target gene, Rab37, primer pairs were a sense primer 5'TCGGCATAGACTTCAGGAACAA3' and an antisense primer 5'GCCAGAAAGGCTAACTCCACAT3', with PCR product of 370 bp. The primers were designed to span at list one intron to avoid the contamination of genomic DNA. Cycling conditions included 95°C for 5 min, followed by 35 cycles at 94 °C for 15 s, 61 °C for 15 s and 72 °C for 30 s.
Genotyping
The 3 Tag SNPs in Rab37 gene were enrolled in this study, which were rs9904078G>A, rs2034310T>C, rs5018106T>C, respectively. The minor allele frequency (MAF) of the SNPs was more than 0.05 in Chinese Han populations. Rs9904078 was located in the promoter region, rs2034310 was located in the exton and caused the amino acid at 218 changed form glutamine to arginine, and rs5018106 was located in the intron region of the gene downstream. According to Haploview software [17], the 3 Tag SNPs were located in the different gene blocks. PCR-restriction fragment length polymorphism (PCR-RFLP) assay were performed to achieve the genotypes of the 3 SNPs. The DNA fragments were amplified in a 20 μL reaction volume containing 100 ng genomic DNA and 0.5 μL 10 μM sense and antisense primers (Table 1), 10 μL PCR Master Mix (Sinobio). The cycling program consisted of 95 °C for 5 min, followed by 30 cycles at 94 °C for 30 s, 57-63 °C for 30 s, 72 °C for 25 s, and then 72 °C for 5 min. The successfully amplified 10 μL PCR products were following the overnight digestion at 37 °C with 3 U restriction endonucleases, MboII, AccII and HaeIII, respectively. The expected digestion fragments correspondent with the genotype were summarized in Table 1.
The designs of PCR-FRLP for genotyping.
| Tag SNP | Sense and antisense primers for PCR | Annealing | Endonuclease | Genotype and its expected band(s) (bp) |
|---|---|---|---|---|
| rs9904078 | 5'TAATTTGCCCGAGTTTGC3' 5'GACAGTAGATGGGTCTTAGGTG3' | 57 °C | MboII | G/G:239 G/A:239+145+94 A/A:145+94 |
| rs2034310 | 3'CCCCAGGACTGAGACTTGC5' 3'AGGGCGACCTCTGCTATG5' | 60 °C | AccII | T/T:301 T/C:301+248+53 C/C:248+53 |
| rs5018106 | 5'GACCATTGACCTATACACAAGCCAG3' 5'GACAGAGTCCCCTAGCAGAGCAC3' | 63 °C | HaeIII | T/T:176+29 T/C:176+99+77+29 C/C:99+77+29 |
Statistical analysis
Differences between ESCC patients and cancer-free controls were evaluated by the Student's t and chi-square tests for continuous and categorical variables, respectively. Hardy-Weinberg equilibrium was applied by a chi-square test to compare the observed genotype frequencies with the expected. A 5% probability level was taken as statistically significant and the odds ratio (OR) was calculated with 95% confidence interval (CI). The statistical analyses were performed using the SPSS software (version 13). The haplotype analysis on the polymorphisms was performed using SHEsis software [18].
Results
The expression of Rab37 mRNA in ESCC cell lines
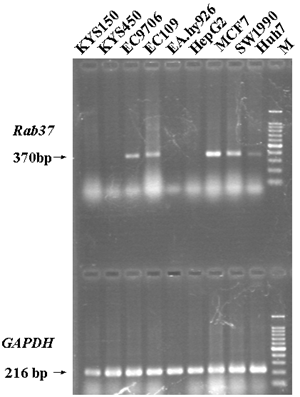
Rab37 was initially thought as a mast cell specific GTPase and then its expression could be limited to some specific cells with secretory functions [2-5]. To our surprise, the expression of Rab37 mRNA was also found in two ESCC cell lines, EC109 and EC9706, but not in KYS150 and KYS450, though the all four cell lines were from the esophageal epithelial cells of ESCC patients [19-21]. The cell-specific expression of Rab37 was also proven by non-ESCC cell lines. The expression of Rab37 mRNA was positive in MCF7, SW1990 and Huh7, but negative in EA.hy926 and HepG2 cells lines (Figure 1).
The expression of Rab37 mRNA in different cell lines by RT-PCR. Lane M, 100bp marker.
Association analysis between the polymorphisms of Rab37 and the risk of ESCC
As the expression of Rab37 mRNA was found in ESCC, we wondered whether its polymorphisms would be associated with ESCC. Thus, genotyping analysis for Tag SNPs of Rab37 was performed in this study. To avoid the population stratification of age and gender, 212 ESCC surgery patients and 213 controls were included in our case-control sample pool. Their ages were ranged from 50 to 61 and the ratio of men to women was about 3 to 1. Furthermore, all ESCC patients were prior verified as Rab37 positive in their surgery tumor tissue samples by RT-PCR. The characteristics of these subjects were summarized in Table 2.
General characteristics of the subjects.
| ESCC patient | Cancer free control | P value | |
|---|---|---|---|
| Total | 212 | 213 | |
| Age (years, mean±SD) | 55.28±2.78 | 55.30±3.19 | 0.613 |
| Gender | |||
| Male (N, %) | 163(76.9) | 157(73.7) | |
| Female (N, %) | 49(23.1) | 56(26.3) | 0.448 |
| TNM Stage | |||
| I (N, %) | 91(42.9) | ||
| IIa (N, %) | 71(33.5) | ||
| IIb (N, %) | 12(5.7) | ||
| III (N, %) | 21(9.9) | ||
| IV (N, %) | 17(8.0) |
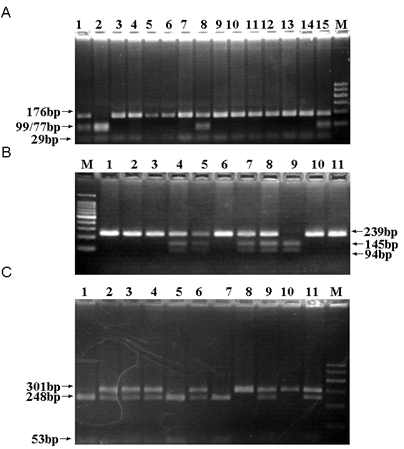
To discriminate Rab37 genotypes at 3 Tag SNP sites, rs9904078, rs2034310, rs5018106, three restriction endonucleases, MboII, AccII and HaeIII, were used respectively. The typical PCR-RFLP detections of the samples were shown in Figure 2. All SNPs were successfully genotyped with the PCR-RFLP method and 10% of the samples were randomly selected for sequencing verification. The results were completely concordant in our experimental system. As Table 3 shown, the observed genotype frequencies for the three polymorphisms were in agreement with that expected under the Hardy-Weinberg equilibrium in the case and control groups (P>0.05), suggesting that the samples in this study were from a genetic equilibrium population without the deviation of the artificial selection. Furthermore, as for our data on the MAF of rs9904078, A allele was 0.154 in patients and 0.124 in control respectively and the MAF of rs9904078 in the Beijing Han Chinese (CHB) population in the database of SNP (dbSNP) is about 0.122-0.167. As for the MAF of rs2034310, T allele was 0.290 in patients and 0.343 in control respectively, which are included in the range of that in CHB (0.221-0.378) in dbSNP. Also, as for the MAF of rs5018106, C allele was 0.113 in patients and 0.124 in control respectively, which are included in the range of that in CHB (0.089-0.195) in dbSNP. All these data indicated that our experimental allele frequencies of 3 Tag SNPs were consistent with the recorded features of CHB in dbSNP.
To evaluate the relationship between the polymorphisms of Rab37 and ESCC, association analyses were applied in this study. As Table 3 shown, any genotypic or allelic frequency was not significantly different between the patients and controls for all 3 SNPs (P>0.05). Furthermore, all SNPs genotypes of every sample were entered into SHEsis software for haplotype analysis. All analyzed haplotypes consisted with 3 SNP sites in the patients did not differ from those in the controls (P>0.05). Those data demonstrated that there was no significant association between the polymorphisms of Rab37 and the risk of ESCC cancer.
The typical PCR-RFLP determination of the samples. The genotypes of rs5018106 (A): T/T (Lane 3-7, 9-14), T/C (Lane 1, 8, 15) and C/C (Lane 2). The genotypes of rs9904078 (B): G/G (Lane 1-3, 6, 10, 11), G/A (Lane 4, 5, 7, 8) and A/A (Lane 9). The genotypes of rs2034310 (C): T/T (Lane 8, 10), T/C (Lane 2-4, 6, 9, 11) and C/C (Lane 1, 5, 7). Lane M, 100bp marker.
Association analysis between the polymorphisms of Rab37 and the risk of ESCC in ESCC patients and cancer free controls.
| Patients | Controls | P value | OR [95%CI] | P value | |
|---|---|---|---|---|---|
| rs9904078 | |||||
| H-W P value | 0.6032 | 0.6583 | |||
| Genotype | N=212 | N=213 | 0.378 | ||
| G/G | 151(71.2%) | 164(77.0%) | 1a | ||
| A/G | 57(26.9%) | 45(21.1%) | 1.38[0.88-2.16] | 0.163 | |
| A/A | 4(1.9%) | 4(1.9%) | 1.09[0.27-4.42] | 0.908 | |
| A/G+A/Ab | 61(28.8%) | 49(23.0%) | 0.175 | 1.35[0.87-2.09] | 0.175 |
| A/G+G/Gc | 208(98.1%) | 209(98.1%) | 0.995 | 1.00[0.25-4.03] | 0.995 |
| Allele | 2N=414 | 2N=426 | 0.223 | ||
| G | 359(84.6%) | 373(87.6%) | 1a | ||
| A | 65(15.4%) | 53(12.4%) | 1.27[0.86-1.88] | 0.223 | |
| rs2034310 | |||||
| H-W P value | 0.5393 | 0.9954 | |||
| Genotype | N=212 | N=213 | 0.227 | ||
| C/C | 105(49.5%) | 92(43.2%) | 1a | ||
| C/T | 91(42.9%) | 96(45.1%) | 0.83[0.56-1.24] | 0.364 | |
| T/T | 16(7.5%) | 25(11.7%) | 0.56[0.28-1.11] | 0.096 | |
| C/T+T/Td | 107(50.5%) | 121(56.8%) | 0.190 | 0.77[0.53-1.14] | 0.190 |
| C/T+C/Ce | 196(92.5%) | 188(88.3%) | 0.144 | 1.63[0.84-3.15] | 0.144 |
| Allele | 2N=414 | 2N=426 | 0.099 | ||
| T | 123(29.0%) | 146(34.3%) | 1a | ||
| C | 301(71.0%) | 280(65.7%) | 1.28[0.95-1.71] | 0.099 | |
| rs5018106 | |||||
| H-W P value | 0.8465 | 0.8518 | |||
| Genotype | N=212 | N=213 | 0.849 | ||
| T/T | 167(78.8%) | 163(76.5%) | 1a | ||
| C/T | 42(19.8%) | 47(22.1%) | 0.87[0.55-1.39] | 0.567 | |
| C/C | 3(1.4%) | 3(1.4%) | 0.98[0.19-4.91] | 0.977 | |
| C/T+C/Cf | 45(21.2%) | 50(23.5%) | 0.578 | 0.88[0.56-1.39] | 0.578 |
| C/T+T/Tg | 209(98.6%) | 210(98.6%) | 0.995 | 1.00[0.20-4.99] | 0.995 |
| Allele | 2N=414 | 2N=426 | 0.614 | ||
| T | 376(88.7%) | 373(87.6%) | 1a | ||
| C | 48(11.3%) | 53(12.4%) | 0.90[0.59-1.36] | 0.614 | |
| Haplotype | 2N=414 | 2N=426 | 0.192 | ||
| A-C-C h | 0.51(0.1%) | 0.33(0.1%) | - | - | - |
| A-C-T | 50.85(12.0%) | 40.84(9.6%) | 0.255 | 1.288 [0.83-1.99] | 0.255 |
| A-T-C h | 3.42(0.8%) | 3.01(0.7%) | - | - | - |
| A-T-T | 10.22(2.4%) | 8.81(2.1%) | 0.734 | 1.171 [0.47-2.91] | 0.734 |
| G-C-C | 12.34(2.9%) | 4.96(1.2%) | 0.071 | 2.551 [0.89-7.30] | 0.071 |
| G-C-T | 237.30(56.0%) | 233.87(54.9) | 0.736 | 1.048 [0.80-1.38] | 0.736 |
| G-T-C | 31.73(7.5%) | 44.70(10.5%) | 0.127 | 0.691 [0.43-1.11] | 0.127 |
| G-T-T | 77.63(18.3%) | 89.48(21.0%) | 0.328 | 0.844 [0.60-1.19] | 0.328 |
OR: Odds Ratio. CI: Confidence Intervals. H-W: Hardy-Weinberg equilibrium. a Reference genotype/allele. b Comparing subjects with A/G+A/A genotypes vs. G/G genotype. c Comparing subjects with A/G+G/G genotypes vs. A/A genotype. d Comparing subjects with C/T+T/T genotypes vs. C/C genotype. e Comparing subjects with C/T+C/C genotypes vs. T/T genotype. f Comparing subjects with C/T+C/C genotypes vs. T/T genotype. g Comparing subjects with C/T+T/T genotypes vs. C/C genotype. h All those frequency < 1 % will be ignored in analysis.
The association analysis between the polymorphisms of Rab37 and the clinic TNM stage of ESCC
In addition, we analyzed the association between the polymorphisms of Rab37 and the clinic TNM stage of ESCC patients in Table 4. The allele and genotype distributions were not significantly different between the stage I-IIa and stage IIb-IV patients (P>0.05), indicating no association between the polymorphisms and the lymphonodus transfer or metastasis of ESCC.
The association analysis between the polymorphisms of Rab37 and the clinic TNM stage of ESCC patients.
| TNM stage | P value | OR [95%CI] | P value | ||
|---|---|---|---|---|---|
| I-IIa | IIb-IV | ||||
| rs2034310 | |||||
| Genotype | N=162 | N=50 | 0.983 | ||
| T/T | 12(7.4%) | 4(8.0%) | 1a | ||
| C/T | 70(43.2%) | 21(42.0%) | 1.11[0.32-3.81] | 0 .867 | |
| C/C | 80(49.4%) | 25(50.0%) | 1.07[0.32-3.60] | 0 .917 | |
| C/T+C/Cb | 150(92.6%) | 46(92.0%) | 0 .890 | 1.09[0.33-3.53] | 0 .890 |
| C/T+T/Tc | 82(50.6%) | 25(50.0%) | 0 .939 | 1.02[0.54-1.93] | 0 .939 |
| Allele | 2N=324 | 2N=100 | 0.998 | ||
| T | 94(29.0%) | 29(29.0%) | 1a | ||
| C | 230(71.0%) | 71(71.0%) | 1.00 [0.61-1.64] | 0.998 | |
| rs5018106 | |||||
| Genotype | N=162 | N=50 | 0.927 | ||
| T/T | 128(79.0%) | 39(78.0%) | 1a | ||
| C/T | 32(19.8%) | 10(20.0%) | 0.98 [0.44-2.16] | 0.950 | |
| C/C | 2(1.2%) | 1(2.0%) | 0.61 [0.05-6.90] | 0.686 | |
| C/T+C/Cd | 34(21.0%) | 11(22.0%) | 0.878 | 0.94[0.44-2.03] | 0.878 |
| C/T+T/Te | 160(98.8%) | 49(98.0%) | 1.000 | 1.63[0.14-18.39] | 1.000 |
| Allele | 2N=324 | 2N=100 | 0.806 | ||
| T | 288(88.9%) | 88(88.0%) | 1a | ||
| C | 36(11.1%) | 12(12.0%) | 0.92 [0.46-1.84] | 0.806 | |
| rs9904078 | |||||
| Genotype | N=162 | N=50 | 0.986 | ||
| G/G | 115(71.0%) | 36(72.0%) | 1a | ||
| A/G | 44(27.2%) | 13(26.0%) | 1.06 [0.51-2.18] | 0.875 | |
| A/A | 3(1.9%) | 1(2.0%) | 0.94 [0.09-9.31] | 0.957 | |
| A/G+A/Af | 47(29.0%) | 14(28.0%) | 0.890 | 1.05[0.52-2.13] | 0.890 |
| A/G+G/Gg | 159(98.1%) | 49(98.0%) | 1.000 | 1.08[0.11-10.64] | 1.000 |
| Allele | 2N=324 | 2N=100 | 0.917 | ||
| G | 274(84.6%) | 85(85.0%) | 1a | ||
| A | 50(15.4%) | 15(15.0%) | 1.03 [0.56-1.93] | 0.917 | |
OR: Odds Ratio. CI: Confidence Intervals. a Reference genotype/allele. b Comparing subjects with C/T+C/C genotypes vs. T/T genotype. c Comparing subjects with C/T+T/T genotypes vs. C/C genotype. d Comparing subjects with C/T+C/C genotypes vs. T/T genotype. e Comparing subjects with C/T+T/T genotypes vs. C/C genotype. f Comparing subjects with A/G+A/A genotypes vs. G/G genotype. g Comparing subjects with A/G+G/G genotypes vs. A/A genotype.
Discussion
To our knowledge, there are no more than 12 researches on Rab37 and no report on the association between Rab37 and ESCC. As previously shown, Rab37 was expressed in some secreting cells and two types of cancer cells [2-7]. Here, Rab37 mRNA was detected in two ESCC cell lines, EC109 and EC9706 [20, 21], established from Chinese patients but not in the other two cell lines, KYS150 and KYS450 [19], established from Japanese sufferers, though none of them were the EA type of esophageal cancer patients. Interestingly, the expression of Rab37 mRNA was different in two hepatic cell lines, HepG2 and Huh7 (Figure 1). Both were sourced from hepatocellular carcinoma patients but HepG2 was established from a Caucasian male while Huh7 was from a Japanese patient [22, 23]. Our data supports the point of view that the expression of Rab37 has obvious cell specificity.
Not only the SNP loci but also the population would greatly contribute to the results of association analysis in case-control studies. Recently, two large-scale genome-wide association studies simultaneously reported the association of some susceptibility loci with the risk of ESCC in Chinese population [24, 25]. But as to C20orf54 gene, the results were inconsistent in the two GWAS studies. Even in their following focused testing, the variation of C20orf54 would be a risk factor for esophageal cancer patients form Shanxi Province (the North of China) but not from Jiangsu Province (the East of China) [26, 27]. In this report, we failed to find the association of the 3 Tag SNPs, rs9904078, rs2034310 and rs5018106, located in Rab37 gene region with the risk of ESCC. Its allele, genotype and haplotype frequencies were not significantly different between the ESCC patients and the cancer-free controls as shown in Table 3. According to the dbSNP, the MAF of rs9904078 in CHB population is about 0.122-0.167 while its MAF in Utah residents with Northern and Western European (CEU) is about 0.5. Our data on the MAF of rs9904078 were consistent with that in CHB and obviously lower than that of CEU. Furthermore, our data on the MAF of rs2034310 were included in the range of that in CHB and obviously higher than that in CEU (0.173-0.183). If the two SNPs, rs9904078 and rs2034310, could be analyzed in non-Chinese populations, the more information on the relationship between Rab37 gene and the risk of ESCC would be further discovered.
We supposed that the cell-specific expression of Rab37 in ESCC might have its specific functions on esophageal cancer aetiology because some cancer-related hot star molecules, such as E-cadherin and integrin, are regulated by endocytic and exocytic pathways [28, 29]. Then the stratification analysis between the polymorphisms of Rab37 and the clinic TNM stage of ESCC patients were performed, but beyond our expectations its polymorphisms were lack of the association with the risk and the lymphonodus transfer or metastasis of ESCC as the data shown in Table 4, which would be different from the inference from NSCLC [7]. Obviously the sample size could impact the stratification analysis of TNM stage so our result might be influenced by the sample size of included ESCC patients. In fact, the sufferer is not suitable for surgery before chemotherapy and/or radiation therapy if he or she is suspected as an ESCC patient with metastasis. And the 50 TNM IIb-IV patients included in our study were just occasionally confirmed during their operations. Thus, it's impossible to collect the enough ESCC patients with advanced TNM stage except the multicenter collaboration. And the further research should be carried out to explore the significance of Rab37 gene expression in ESCC
Conclusion
We found that Rab37 mRNA could be specifically detected in some ESCC cell lines, but its genetic polymorphisms were not associated with ESCC in a Chinese Han population residing in Chongqing City of southwest China.
Abbreviations
ESCC: esophageal squamous cell carcinoma; RT-PCR: reverse transcription-polymerase chain reaction; RCC: renal cell carcinoma; NSCLC: non-small cell lung cancer; TSG: tumor suppressor gene; EA: esophageal adenocarcinoma; SNPs: single nuclear polymorphisms; MAF: minor allele frequency; RFLP: restriction fragment length polymorphism; OR: odds ratio; CI: confidence interval; CHB: Beijing Han Chinese; dbSNP: database of SNP.
Acknowledgements
This work was partially supported by grants 31100131 from the National Natural Science Foundation of China and 2010XQN07 from the Innovational Foundation for Youth of Third Military Medical University.
Competing Interests
The authors declare that they have no conflict of interest.
References
1. Fukuda M. Distinct Rab binding specificity of Rim1, Rim2, rabphilin, and Noc2. Identification of a critical determinant of Rab3A/Rab27A recognition by Rim2. J Biol Chem. 2003;278:15373-80
2. Masuda ES, Luo Y, Young C. et al. Rab37 is a novel mast cell specific GTPase localized to secretory granules. FEBS Lett. 2000;470:61-4
3. Brunner Y, Couté Y, Iezzi M. et al. Proteomics analysis of insulin secretory granules. Mol Cell Proteomics. 2007;6:1007-17
4. Tsuboi T, Fukuda M. Rab3A and Rab27A cooperatively regulate the docking step of dense-core vesicle exocytosis in PC12 cells. J Cell Sci. 2006;119:2196-203
5. Mori R, Ikematsu K, Kitaguchi T. et al. Release of TNF-α from macrophages is mediated by small GTPase Rab37. Eur J Immunol. 2011;41:3230-9
6. Dobashi S, Katagiri T, Hirota E. et al. Involvement of TMEM22 overexpression in the growth of renal cell carcinoma cells. Oncol Rep. 2009;21:305-12
7. Wu CY, Tseng RC, Hsu HS. et al. Frequent down-regulation of hRAB37 in metastatic tumor by genetic and epigenetic mechanisms in lung cancer. Lung Cancer. 2009;63:360-7
8. Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137-50
9. Hoffman AE, Zheng T, Yi C. et al. MicroRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysis. Cancer Res. 2009;69:5970-7
10. Pickens A, Orringer MB. Geographical distribution and racial disparity in esophageal cancer. Ann Thorac Surg. 2003;76:S1367-9
11. Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241-52
12. Yu MC, Garabrant DH, Peters JM. et al. Tobacco alcohol diet occupation and carcinoma of the esophagus. Cancer Res. 1988;48:3843-8
13. Yang X, Xiong G, Chen X. et al. Polymorphisms of survivin promoter are associated with risk of esophageal squamous cell carcinoma. J Cancer Res Clin Oncol. 2009;135:1341-9
14. Xiong G, Guo H, Wang K. et al. Polymorphisms of decoy receptor 3 are associated with risk of esophageal squamous cell carcinoma in Chinese Han. Tumour Biol. 2010;31:443-9
15. Wang K, Guo H, Hu H. et al. A functional variation in pre-microRNA-196a is associated with susceptibility of esophageal squamous cell carcinoma risk in Chinese Han. Biomarkers. 2010;15:614-8
16. Guo H, Wang K, Xiong G. et al. A functional varient in microRNA-146a is associated with risk of esophageal squamous cell carcinoma in Chinese Han. Fam Cancer. 2010;9:599-603
17. Barrett JC, Fry B, Maller J. et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263-5
18. Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97-8
19. Shimada Y, Imamura M, Wagata T. et al. Characterization of 21 Newly Established Esophageal Cancer Cell Lines. Cancer. 1992;69:277-84
20. Wang HT, Kong JP, Ding F. et al. Analysis of gene expression profile induced by EMP-1 in esophageal cancer cells using cDNA Microarray. World J Gastroenterol. 2003;9:392-8
21. Doki Y, Imoto M, Han EK. et al. Increased expression of the P27KIP1 protein in human esophageal cancer cell lines that over-express cyclin D1. Carcinogenesis. 1997;18:1139-48
22. Diamond L, Kruszewski F, Aden DP. et al. Metabolic activation of benzo[a]pyrene by a human hepatoma cell line. Carcinogenesis. 1980;1:871-5
23. Nakabayashi H, Taketa K, Yamane T. et al. Phenotypical stability of a human hepatoma cell line, HuH-7, in long-term culture with chemically defined medium. Gann. 1984;75:151-8
24. Wang LD, Zhou FY, Li XM. et al. Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nat Genet. 2010;42:759-63
25. Abnet CC, Freedman ND, Hu N. et al. A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nat Genet. 2010;42:764-7
26. Gu H, Ding G, Zhang W. et al. Replication study of PLCE1 and C20orf54 polymorphism and risk of esophageal cancer in a Chinese population. Mol Biol Rep. 2012;39:9105-11
27. Ji A, Wang J, Yang J. et al. Functional SNPs in human C20orf54 gene influence susceptibility to esophageal squamous cell carcinoma. Asian Pac J Cancer Prev. 2011;12:3207-12
28. Bryant DM, Stow JL. The ins and outs of E-cadherin trafficking. Trends Cell Biol. 2004;14:427-34
29. Pellinen T, Ivaska J. Integrin traffic. J Cell Sci. 2006;119:3723-31
Author contact
![]() Corresponding author: Prof. Yun Bai, Department of Medical Genetics, Third Military Medical University, Chongqing 400038, People's Republic of China. Tel: +86 23 68752258; E-mail: baiyungenecom or Prof. Kang Yang, Department of Thoracic and Cardiac Surgery, Southwest Hospital, Third Military Medical University, Chongqing 400038, People's Republic of China. Tel: +86 23 68754683; E-mail: kangyangedu.cn.
Corresponding author: Prof. Yun Bai, Department of Medical Genetics, Third Military Medical University, Chongqing 400038, People's Republic of China. Tel: +86 23 68752258; E-mail: baiyungenecom or Prof. Kang Yang, Department of Thoracic and Cardiac Surgery, Southwest Hospital, Third Military Medical University, Chongqing 400038, People's Republic of China. Tel: +86 23 68754683; E-mail: kangyangedu.cn.

 Global reach, higher impact
Global reach, higher impact