3.2
Impact Factor
ISSN: 1449-1907
Int J Med Sci 2013; 10(3):206-221. doi:10.7150/ijms.5529 This issue Cite
Review
Systematic Reviews of Animal Models: Methodology versus Epistemology
Americans For Medical Advancement, 2251 Refugio Rd, Goleta, CA 93117, USA.
* The authors contributed equally to the article.
Received 2012-11-12; Accepted 2012-12-30; Published 2013-1-11
Abstract
Systematic reviews are currently favored methods of evaluating research in order to reach conclusions regarding medical practice. The need for such reviews is necessitated by the fact that no research is perfect and experts are prone to bias. By combining many studies that fulfill specific criteria, one hopes that the strengths can be multiplied and thus reliable conclusions attained. Potential flaws in this process include the assumptions that underlie the research under examination. If the assumptions, or axioms, upon which the research studies are based, are untenable either scientifically or logically, then the results must be highly suspect regardless of the otherwise high quality of the studies or the systematic reviews. We outline recent criticisms of animal-based research, namely that animal models are failing to predict human responses. It is this failure that is purportedly being corrected via systematic reviews. We then examine the assumption that animal models can predict human outcomes to perturbations such as disease or drugs, even under the best of circumstances. We examine the use of animal models in light of empirical evidence comparing human outcomes to those from animal models, complexity theory, and evolutionary biology. We conclude that even if legitimate criticisms of animal models were addressed, through standardization of protocols and systematic reviews, the animal model would still fail as a predictive modality for human response to drugs and disease. Therefore, systematic reviews and meta-analyses of animal-based research are poor tools for attempting to reach conclusions regarding human interventions.
Keywords: Systematic reviews, axiom, biological complexity, evolution, animal models.
Introduction
Review articles in the scientific literature can be classified as a general review article, a systematic review (SR), or meta-analysis (MA). The purpose of a review article is to provide readers with a summary of published research in a particular field. Reviews usually focus on areas of progress over the recent past, for example five years. A general review article attempts to summarize all the relevant, published literature and provide some analysis of the controversial areas of the field or topic. In addition, it may suggest some novel ways to advance the field further [1]. Such review articles provide a concise analysis of a large body of literature and hence are important for readers from a variety of fields. Articles in PubMed, for example, can be searched based on whether they are classified as review articles.
In contrast, SRs seek to be more rigorous and comprehensive in addition to providing an opinion about outcomes or practice. For example, in medical science, SRs are used in hopes of ascertaining whether treatment A is superior to treatment B. Why is such an analysis necessary? Unfortunately, few research protocols are perfect so there may be controversy surrounding treatment options even after numerous studies. Therefore, combining studies and analyzing them may be useful. However, there is another reason explained by Greenhalgh: “Experts, who have been steeped in a subject for years and know what the answer 'ought' to be, are less able to produce an objective review of the literature in their subject than non-experts. This would be of little consequence if experts' opinions could be relied on to be congruent with the results of independent systematic reviews, but they cannot” [2]. One of the premises upon which the practice of SRs is based, is the inability of informed scientists to evaluate, without bias, the controversies in their own field. This is a reflection of human nature and is unlikely to change anytime in the near future [3]. A SR requires clearly stated objectives and rigorous criteria for what studies can and cannot be included, should be reproducible, include all relevant studies, seek to detect bias, and attempt to make determinations [4, 5]. SRs are acknowledged as being an integral component of evidence based medicine, where the goal is to analyze the evidence gained from the best scientific studies that qualify for consideration in order to make a determination regarding clinical intervention. The SR is thus “the conscientious, explicit, judicious use of current best evidence in making decisions about the care of individual patients” [6].
The term meta-analysis was coined in 1980 by Smith, Glass and Miller and involves a statistical analysis of the topic of a SR. A MA can be thought of as a quantitative SR [7]. Greenhalgh stated: “A meta-analysis is a mathematical synthesis of the results of two or more primary studies that addressed the same hypothesis in the same way” [2].
While the purpose of any scientific literature review is to summarize and evaluate relevant articles in a scholarly and rigorous manner, the review must also consider relevant research in other disciplines of science—consilience—as well as the scientific underpinnings of the topic under consideration. For example, any SR of research articles regarding acupuncture should take place in light of the fact that no mechanisms have been discovered that would allow scientists to expect success from using acupuncture in order to alleviate objective pathology [8, 9]. In contrast, the Germ Theory of Disease supports a SR of the efficacy of antibacterial use for preventing complications from, or shortening the course of, ear infections in children. An example of this concept comes from oncological surgeon David Gorski who criticized the National Center for Complementary and Alternative Medicine (NCCAM) for spending resources to study: “treatment modalities that are inherently unscientific, being as they are based on prescientific or demonstrably incorrect understandings of human physiology and disease” [10]. An example of knowledge from other fields of science affecting how a SR might be conducted can be found in homeopathy. Knowledge from chemistry and physics vis-à-vis how to apply Avogadro's number when calculating dilutions, should inform scientists seeking to evaluate homeopathy by conducting a SR [11].
Finally, the fact that conclusions drawn from SRs and MAs have been shown to be wrong should also be considered when evaluating a treatment or other practice being evaluated by a SR or MA. For example, a meta-analysis by the Cochrane Group reported that albumin increased deaths in critically ill patients [12]. However, a large randomized study in Australia later revealed no such effects [13]. In summary, SRs and MAs are a valuable tool in assessing what is currently known regarding a subject but, like any tool, can fail.
Systematic reviews and standardization of animal model protocols
Because nonhuman animal models (hereafter referred to as animal models or animals) have on multiple occasions been unsuccessful in predicting human response to drugs and disease (we will address this claim in depth), many have called for SRs in order to improve the models [14-24]. An example of this predicament would be the animal models used to determine which drugs to develop in an attempt to diminish neurological damage from ischemia events of the central nervous system (CNS) [17, 25-30]. By analyzing animal-based research with SRs, flaws in the methodology would also become apparent thus leading to eventual standardization of such studies. This would ostensibly also lead to better predictive values for humans (see table 1 for calculating such values). Bracken supports this, stating:
One reason why animal experiments often do not translate into replications in human trials or into cancer chemoprevention is that many animal experiments are poorly designed, conducted and analyzed. Another possible contribution to failure to replicate the results of animal research in humans is that reviews and summaries of evidence from animal research are methodologically inadequate [18].
Binary classification and formulas for calculating predictive values of modalities such as animal-based research.
| Gold Standard | |||
|---|---|---|---|
| GS+ | GS- | ||
| Test | T+ | TP | FP |
| T- | FN | TN | |
| Sensitivity = TP/(TP+FN) | |||
| Specificity = TN/(FP+TN) | |||
| Positive Predictive Value = TP/(TP+FP) | |||
| Negative Predictive Value = TN/(FN+TN) | |||
| T- = Test negative T+ = Test positive FP = False positive TP = True positive FN = False negative TN = True negative GS- = Gold standard negative GS+ = Gold standard positive | |||
Further evidence that SRs are expected to transform the predictive value of animal-based research comes in the form of the 1st International Symposium and Workshop on Systematic Reviews in Laboratory Animal Science that was held at the Radboud University Nijmegen Medical Centre on February 9-10, 2012. The workshop celebrated “5 years of the 3R [the 3R here refers to Reduce, Refine, and Replace animals used in research] Research Centre (3RRC) and stimulating an international discussion and collaboration between animal and clinical researchers on Systematic Reviews (SRs) of animal studies”[31]. Malcolm Macleod, the keynote speaker, discussed, “The transforming potential of the systematic evaluation of laboratory research.” The brochure for the conference stated:
. . . the use of SRs for the optimisation of animal testing is still rare which can lead to waste in funding and harm to patients and research volunteers. The 3RRC encourages the use of SRs in animal studies as they improve scientific quality, lead to implementation of the 3Rs principles, improve translational research and help in determining the value of animal studies to human health [31].
There are several claims here for the value of SRs. While we do not dispute the value of SRs to improve the quality of research and perhaps increase acceptance of the 3Rs, we strongly contest the notion that SRs will allow scientists to develop animal models that are predictive modalities for human responses to drugs and disease. Claims such as those above by Bracken and the organizers of the Symposium (and more we will cite below) regarding the benefit of using animal models in translational research however, directly assumes predictive ability. We will discuss this further when describing table 2.
There are methodological problems in current animal-based research. Pound et al. [32] highlighted some of the potential flaws when using animal models, including:
- Variations in drug dosing schedules and regimens that are of uncertain relevance to the human condition.
- Variability in the way animals are selected for study, methods of randomization, choice of comparison therapy (none, placebo, vehicle), and reporting of loss to follow up.
- Small experimental groups with inadequate power, simplistic statistical analysis that does not account for potential confounding, and failure to follow intention to treat principles.
- Nuances in laboratory technique that may influence results may be neither recognized nor reported, e.g. methods for blinding investigators.
- Selection of a variety of outcome measures, which may be disease surrogates or precursors and which are of uncertain relevance to the human clinical condition.
- Length of follow up before determination of disease outcome varies and may not correspond to disease latency in humans [32].
Hooijmans et al [14] have called for a gold standard for research involving animals that includes stating the specifics regarding housing, species, randomization, cage size and bedding among other parameters. Other checklists and suggestions aimed toward improving standardizations have also been published [21, 33-38]. Note that even here however, Hooijmans et al link standardization to prediction of human response stating: “In addition, an improved experimental design contributes to a better translation to the clinic and increases patient safety” [14]. Many reviews and opinions have echoed the above reasons for translational failure or predictive failure and have suggested ways to improve the likelihood of successfully predicting human responses to drugs and disease. The ARRIVE (Animals in Research: Reporting In Vivo Experiments) Guidelines for Reporting Animal Research [39] consist of a 20 item checklist containing:
the minimum information that all scientific publications reporting research using animals should include, such as the number and specific characteristics of animals used (including species, strain, sex, and genetic background); details of housing and husbandry; and the experimental, statistical, and analytical methods (including details of methods used to reduce bias such as randomization and blinding) [39].
Another example of such an effort is the CAMARADES group (the Collaborative Approach to Meta-Analysis and Review of Animal Data in Experimental Studies). For example, the CAMARADES group identified significant sources of bias in a sample of almost 5,000 animal studies. These shortcomings included a frequent lack of blinding, randomization, and sample size calculation, in addition to overstatement of treatment efficacy due to unpublished studies.
While some scientists are more modest in their claims for the value of SRs and the standardization of protocols, clearly there are high hopes for what SRs can accomplish regarding the predictive value of animal models. We will now examine, in more depth, the reasons for the above concerns regarding the predictive value of animal models.
Prediction in science
The use of animals in science and research can be categorized per table 2 [40]. While all uses of sentient animals are cause for ethical concern [41-43], the use of animal models to predict human response to drugs and disease appears to be the main focus of the scientific community when attempting to justify animal use to society [44-52] [[53] p3]. This is consistent with Giles, writing in Nature, who stated:
In the contentious world of animal research, one question surfaces time and again: how useful are animal experiments as a way to prepare for trials of medical treatments in humans? The issue is crucial, as public opinion is behind animal research only if it helps develop better drugs. Consequently, scientists defending animal experiments insist they are essential for safe clinical trials, whereas animal-rights activists vehemently maintain that they are useless [54].
Statements from advocates for animal-based research acknowledge the importance society places on animal models being able to predict human response to drugs and disease. For example, Cheng stated: “Animal tests are necessary for some research, such as testing drugs for toxicity. It would be, in my opinion, improper to release drugs for human use without animal testing” [55]. Heywood likewise stated: “Animal studies fall into two main categories: predictive evaluations of new compounds and their incorporation into schemes designed to help lessen or clarify a recognised hazard” [56]. Vassar agrees, stating: “Chronic dosing in mice and monkeys is necessary to show the efficacy and safety of the antibody before it's taken into humans” [51]. The Council for International Organizations of Medical Sciences implies prediction when they state: “clinical testing must be preceded by adequate laboratory or animal experimentation to demonstrate a reasonable probability of success without undue risk” [45]. Rudczynski wrote: “the basic research model used by Yale University and its peer institutions is scientifically valid and predictive of human disease” [57]. (Emphasis added.) Such statements could be easily multiplied. The animal-based research community clearly stresses the importance and validity of using animals to predict human response to drugs and disease.
Nine categories of animal use in science and research.
| 1. Animals are used as predictive models of humans for research into such diseases as cancer and AIDS. |
| 2. Animals are used as predictive models of humans for testing drugs or other chemicals. |
| 3. Animals are used as “spare parts”, such as when a person receives an aortic valve from a pig. |
| 4. Animals are used as bioreactors or factories, such as for the production of insulin or monoclonal antibodies, or to maintain the supply of a virus. |
| 5. Animals and animal tissues are used to study basic physiological principles. |
| 6. Animals are used in education to educate and train medical students and to teach basic principles of anatomy in high school biology classes. |
| 7. Animals are used as a modality for ideas or as a heuristic device, which is a component of basic science research. |
| 8. Animals are used in research designed to benefit other animals of the same species or breed. |
| 9. Animals are used in research in order to gain knowledge for knowledge sake. |
The above claims are, however, in direct opposition to those advocating for SRs in order to improve the predictive ability of animal-based research. Before we survey the literature for empirical confirmation and present views of other scientists that strongly disagree with the above, we need to first define the term predict and refresh the reader's memory of how it is used in science.
Predict can be used in essentially two ways when discussing science. First, scientists develop hypotheses, which generate predictions that can then be tested. Several confirmations of the hypothesis, by predictions that are found to be true, strengthen the hypothesis while one failed prediction may neccesitate revising the hypothesis or even destroy it altogether. This is standard science based on the hypothetico-deductive method and we have no issues with using the term predict in this manner. Animal use involving categories 5, 7, and 9 in table 2 would employ this use of predict.
The second manner predict is used is when discussing the predictive value of a modality or practice. Such is the case with categories 1 and 2 in table 2. An example outside of biomedical science would be when Italian geologists were asked whether a series of small quakes in the area meant that residents should evacuate their houses because a major earthquake was likely forthcoming. The geologists stated that a major earthquake was unlikely and this was consistent with current knowledge of earthquakes. Nevertheless, the Italian legal system convicted the scientists on charges that essentially said they were negligent in failing to warn the residents to evacuate [58]. This was a cause for concern in the scientific community as an analysis revealed that small quakes forecast a major quake only 2% of the time [59, 60]. Clearly, a practice or modality that correctly calculates the answer only 2% of the time does not qualify as predictive. Exactly what percentage is necessary to qualify will vary with the field of study. Finding a method that will result in the correct answer 51% in the field of gambling, in blackjack for example, would be very productive and probably qualify as meeting the criteria for being a predictive practice. Using instruments to fly an aircraft on the other hand, requires that the instruments correctly communicate the exact location of the aircraft 100% of the time. While medical science does not require predictive values of 100%, it does require very high values. Tests that correlate with reality even 70% of the time are not very useful.
Just as important as what the word predict means in terms of predictive value and how PPV and NPV are calculated, is what does not constitute predictive value. For example, a single example of correlation does not qualify a model as predictive or indicate a high PPV or NPV. A modality or practice must be evaluated based on its history of correlating with reality. Cherry-picking examples is not allowed. Moreover, one must be very precise when defining what is being evaluated for predictive value. If one wishes to evaluate animal models in general then all the wrong answers by all species must be included in the calculation as well as all the correct answers. If one is calculating PPV for a specific animal model, say using beagles in hepatotoxicity testing, then all correct and incorrect answers for beagles should be included but not outcomes from different species or even different breeds.
With that background we can now evaluate the claims of animal models being, or not being, predictive for human response to drugs and disease.
Animals as predictive models
Empirical evidence
The assumption that animal models are predictive of human outcome is foundational for much of their use in biomedical research and for justifying animal-based research in general. Whether this assumption is true is a separate issue from that of methodology and study design although methodology may influence predictive value. The prevailing view within the animal model community among those calling for standardization and SRs, per above, is that animal models would perform better, meaning they would have a higher PPV and NPV for humans, if researchers adhered to strict criteria with respect to study design and methodology [61]. It is important to note that the potential validity of the animal model per se for predicting human response to drugs and disease is not questioned, at least in most of the literature that addresses SRs and standardization. We acknowledge that animals can successfully be used in categories 3-9 in table 2 and that SRs could positively impact on such use and that some calling for SRs and standardization advocate for such on this basis. However, it appears that the main emphasis among those calling for SRs and standardization is to improve predictive value. Therefore we consider it appropriate to explore whether a proper understanding of evolutionary biology and complexity science allows for the use of one species to predict responses to drugs and diseases for another, even under ideal circumstances [40, 41, 62-66]. SRs require the practice under study to be scientifically tenable. If the practice per se is not viable, then SRs will be of little value. We will now present the empirical evidence and later seek to place it within the context of complexity science and evolutionary biology.
Empirical evidence regarding the predictive value of animal models comes in the form of research amenable to quantification via table 1 and examples of multiple failures over many years in the same subject. Examples of the latter would include the search for a vaccine against HIV and neuroprotective drugs. Approximately 100 vaccines have been shown effective against an HIV-like virus in animal models, however, none have prevented HIV in humans [67, 68]. Even if an HIV vaccine came from animal-based research tomorrow, the animal model per se would not be predictive for humans as the PPV would be somewhere in the 0.01 area. Likewise, up to one-thousand drugs have been shown effective for neuroprotection in animal models but none have been effective for humans [23-25, 29, 38, 61, 69-71]. The predictive value is again minimal even if a successful drug is currently in development. The animal model has failed as a modality for predicting neuroprotection. Along the same lines, of twenty two drugs tested on animals and shown to be therapeutic in spinal cord injury, none were effective in humans [72]. As we are attempting to prove that animal models are not predictive such examples are important. Relatively few failures can disqualify a practice from being of predictive value while proving the opposite requires a large number of successes.
The success of the animal model in basic research can also be questioned based on the fact that, according to one report, only 0.004% of basic research papers in leading journals led to a new class of drugs [41, 73] and the fact that the success rate for target identification is similarly dismal [74-78]. For example, in part because the targets derived from animal models are not predictive for humans, the percentage of new drugs in development, after initial evaluation, that ultimately make it to market is somewhere in the area of 0.0002% [79, 80]. We acknowledge that the goals of basic research differ from the goals of applied research where predictive values are most often evaluated. However, because of funding challenges, research that would have historically been considered basic is now being promoted as applied and hence should be judged accordingly [41].
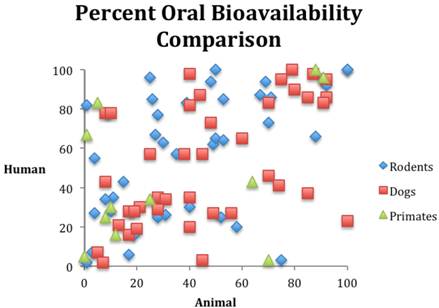
The empirical evidence from research outcomes quantifiable by the calculations in table 1 also supports our position that animal models cannot currently predict human response. Consider the following. In 1962, Litchfield [81] studied rats, dogs, and humans in order to evaluate responses to six drugs. The rat model demonstrated a PPV of 0.49 while the dog model demonstrated a PPV of 0.55. A PPV around 0.5 is not sufficient to qualify a modality as predictive in medical science. It is what one would expect from tossing a coin. Medical science demands values of 0.8 or higher if the modality is to be used for anything that will intersect with patient care. (Drug development is a clear example of a product or modality intersecting with patient care.) A similar study reported in 1990, examined six drugs in animal models, the side effects of which were already known from human data. The study found that at least one species demonstrated 22 side effects, but the models incorrectly identified 48 side effects that did not occur in humans, while missing 20 side effects that did occur in humans. This translates to a PPV of 0.31 [[82] p73]. A similar study - reported in 1990 - examined drugs abandoned during clinical trials secondary to toxicity. In 16 out of 24 cases, the toxicity had no correlation in animal models [[83] 49-56]. A 1994 study revealed that only six of 114 drug toxicities had animal correlates [[84] p57-67]. While the data do not allow the calculations in table 1 to be made, obviously these numbers fall far short of qualifying as a predictive medical modality or test. Likewise, figure 1 illustrates the random nature of bioavailability correlation among species. These examples could be easily multiplied (for example, see [85] [ [86] p67-74] [38, 87-93]). Moreover, in 1995, Lin compared pharmacologically important parameters in different species and pointed out that many examples of animal models predicting human response were in fact retrospective and hence not predictive at all [94].
We acknowledge that the empirical evidence could be interpreted in two ways. First, the animal model per se is simply not predictive of human response to drugs and disease. (For more on the failure of animal models of human disease to correlate with humans, see [62, 64-66, 96-100].) Second, perhaps the proposed SRs and standardization will allow for correction of methodological problems that have resulted in animal models failing to be of predictive value. Perhaps the problem is confined to methodology. In light of this dichotomy, the following questions must be addressed: Is there an all-encompassing explanation for the failure of animal models to be of predictive value regardless of methodology? Is there a theory or law in science that explains the empirical evidence we presented? We propose that the fact that all animals are examples of evolved complex systems constitutes a scientific theory explaining why animal models fail to be predictive modalities for human response to drugs and disease. In addition, this theory requires us to question whether an animal model will ever be a predictive modality for humans at the level of organization where disease and drug response occurs, regardless of methodological improvements.
Comparison of oral bioavailability among three species. Data from reference [95].
Evolved complex systems
Science as a discipline can arguably be dated to Newton and Descartes, both of whom accepted a mechanistic, deterministic universe amenable to study by reductionism [101, 102]. Because the systems under examination at that time were simple systems that were no more than the sum of their parts, exhibited predictable behavior with few interactions and feedback loops, and hence could be intuitively understood, linear cause and effect relationships were the order of the day. Because of the nature of the universe, such systems are amenable to laws while complex systems are usually described using statistics. Hence biological complex systems are more likely to be described by theories than laws [103-105]. Moreover, outcomes are usually described as involving a causal chain as opposed to a linear cause and effect relationship [105].
Ecosystems, climate, financial markets, and the US power grids are examples of complex systems, while humans and animals are examples of evolved complex systems. Reductionism has been of value in the study of complex systems but because of the nature of complex systems, reductionism alone is inadequate to fully describe the system [106-108]. Van Regenmortel states:
The reductionist method of dissecting biological systems into their constituent parts has been effective in explaining the chemical basis of numerous living processes. However, many biologists now realize that this approach has reached its limit. Biological systems are extremely complex and have emergent properties that cannot be explained, or even predicted, by studying their individual parts. The reductionist approach—although successful in the early days of molecular biology— underestimates this complexity and therefore has an increasingly detrimental influence on many areas of biomedical research, including drug discovery and vaccine development [109].
Complex systems have very specific characteristics that influence the ability of one complex system to predict the response of another [102, 106, 107, 109-127].
- Complex systems are more than the sum of their parts, thus reductionism will yield an incomplete analysis of a complex system. As animal modeling is based in large part on reductionism [65, 105, 109, 120, 125, 128-132], this portends problems.
- Complex systems exhibit emergence, meaning that new properties of a complex system arise from the interactions of the parts. These new properties cannot be determined even in light of full knowledge of the component parts, thus compromising reductionism even further.
- Complex systems are resistant to changes and exhibit redundancy in their components. This again complicates extrapolation between complex systems.
- Complex systems exhibit self-organization.
- Complex systems demonstrate responses to perturbations that are nonlinear.
- Complex systems are very dependent upon initial conditions (for example, genetic make-up). For example, strains of mice have been noted to respond very differently to gene deletion [133, 134] and groups of humans, such as sexes [135-140] and ethnic groups [141-149], respond differently to drugs and disease. Monozygotic twins have also been discovered to respond differently to perturbations because of small differences in genetic make-up [150-154].
- Complex systems are composed of many components, which can be grouped into modules that interact with each other.
- Complex systems have hierarchal levels of organization (different levels can even respond oppositely to the same perturbation).
- Complex systems have feedback loops.
- Complex systems interact with their environment—are dynamic.
- Complex systems are nonsimulable [155-158].
Koch describes the problems of studying complex systems:
Such systems [like the human brain] are characterized by large numbers of highly heterogeneous components, be they genes, proteins, or cells. These components interact causally in myriad ways across a very large spectrum of space-time, from nanometers to meters and from microseconds to years. A complete understanding of these systems demands that a large fraction of these interactions be experimentally or computationally probed. This is very difficult. . . . fields as diverse as neuroscience and cancer biology have proven resistant to facile predictions about imminent practical applications. Improved technologies for observing and probing biological systems has only led to discoveries of further levels of complexity that need to be dealt with. This process has not yet run its course. We are far away from understanding cell biology, genomes, or brains, and turning this understanding into practical knowledge [159].
In summary, complex systems are very different from the simple systems described so well by Newtonian physics and which are routinely studied by reductionism. Complex systems are best described by partial differential equations and many of the values of the variables are unknown. Hence predicting intra-complex system response is difficult and predicting inter-complex system response is essentially impossible at higher levels of organization.
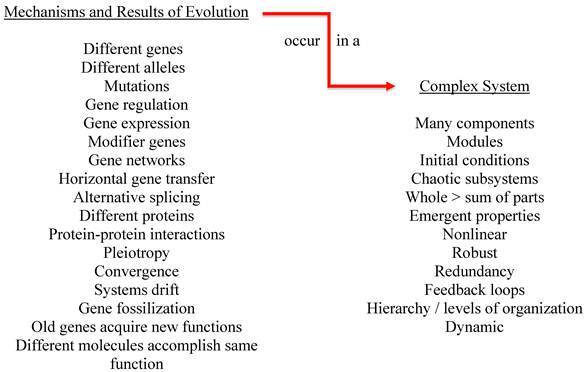
The fact that the complex systems under study have evolved is also significant (see figure 2). While all of the characteristics of a complex system influence inter-system extrapolation, we will illustrate the importance of evolution on just one characteristic—initial conditions. Changes in initial conditions can produce very different outcomes to the same perturbation. Evolution has used numerous mechanisms to match species to niche and all of these mechanisms affect initial conditions. Even among humans, very small differences in genetic makeup can result in dramatically different outcomes to perturbations such as drugs and disease. For example, copy number variants (CNVs) in monozygotic twins can influence outcomes [150]. CNVs have also been shown to influence viral load in HIV patients [160]. Single nucleotide polymorphisms (SNPs) among family members and/or other humans [161-163], pleiotropy [164], alternative splicing [165], the fact that different genes and molecules can accomplish the same purpose, and that the same gene can be used for different purposes [166] all influence response to drugs and disease. Changes in initial conditions such as the presence of different alleles, SNPs, CNVs and so forth negate the similarities between complex systems in terms of predicting response to perturbations that occur at higher levels of organization such as where drug and disease response occurs.
The reality is even more complicated however, as gene regulation and expression account for the major changes in evolution [167, 168]. Theoretically, by varying the regulation and expression of the same genes, a new species could evolve with the same structural genes of its ancestor. Gene expression varies greatly in humans [169-172] and in animals [173-176]. Somel et al. studied gene expression in the brains of humans, chimpanzees, and macaques and discovered accelerated evolution of gene expression in the human prefrontal cortex [177] thus casting doubt on the ability to extrapolate inter-species research for that area. Puente et al discovered at least twenty genes implicated in human cancers that differ significantly from chimpanzees [178]. In addition, chimpanzees are essentially immune to HIV, hepatitis B, and common malaria and they respond differently to other human pathogens [179-182].H According to Caldwell, “It has been obvious for some time that there is generally no evolutionary basis behind the particular-metabolizing ability of a particular species. Indeed, among rodents and primates, zoologically closely related species exhibit markedly different patterns of metabolism” [183]. Festing stated: “There is substantial genetic variation in the response of laboratory rats to xenobiotics, and this variation has important implications for toxicologic research and screening.” Festing goes on to describe a study that reported on “rat” articles published in the journal Toxicology and Applied Pharmacology from 1979 to 1999. In a majority of the articles, the authors did not specify which rat strain was being used [184]. The above has profound consequences for using animal models to predict human response to drugs and disease.
Evolution acts on complex systems.
It is important to note here that many of the scientists quoted above do not take the position that animal models will never be predictive modalities. While we do not want to speculate as to their reasons, we must point out that the fact that animals and humans are evolved complex systems that are differently complex and this leads us to our conclusion that animal models will fail as predictive modalities. The fact, and implications, of models as differently complex is not addressed by most animal modelers quoted above and we suspect this may, in part, explain their position.
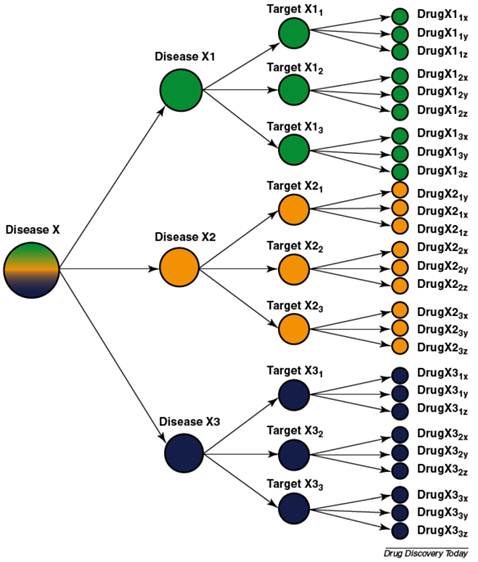
This brings us to the logical conclusion of our animals as evolved complex systems argument. It is also perhaps our best reason against expecting animal models to ever be capable of predicting human response to drugs and disease: the concept of personalized medicine. Personalized medicine is perhaps best illustrated by Allen Roses, then-worldwide vice-president of genetics at GlaxoSmithKline (GSK), who stated: “The vast majority of drugs - more than 90% - only work in 30 or 50% of the people” [185]. Most drugs have an efficacy rate of 50% or lower. Physicians have long recognized intra-species variation in response to drugs and disease [186, 187]. It is now understood that the variations in response are caused by variations in the genome (see tables 3 and 4) including epigenetic changes. For example, because of differences in genes, like SNPs, some children are not protected by a vaccine [162, 163]. King states: “between 5 and 20 per cent of people vaccinated against hepatitis B, and between 2 and 10 per cent of those vaccinated against measles, will not be protected if they ever encounter these viruses” [163]. In the future, such children may be able to receive a personalized vaccine. Personalized medicine will result in medical practice resembling the outline in figure 3 whereas today medical practice is more often “one size fits all.” The fact there is such variation among humans and that this variation causes so much concern [188-197] should cast doubts on the ability of another species to predict human response to drugs and disease [63, 65].
Also illustrative of the problems of extrapolation between complex systems, and in line with the basis for personalized medicine, is the fact that the sexes respond differently to drugs and diseases [135-140, 198], as do ethnic groups [141-149]. Moreover, monozygotic twins respond differently to drugs and disease [74, 199-204]. If monozygotic twins respond differently to perturbations such as drugs and disease, then expecting even genetically modified animals to be of predictive value seems naïve. Indeed genetically modified animals have failed to be of predictive value [74, 199-204]. (For more on personalized medicine see [63, 205, 206].)
The most significant genetic predictors of drug response [208].
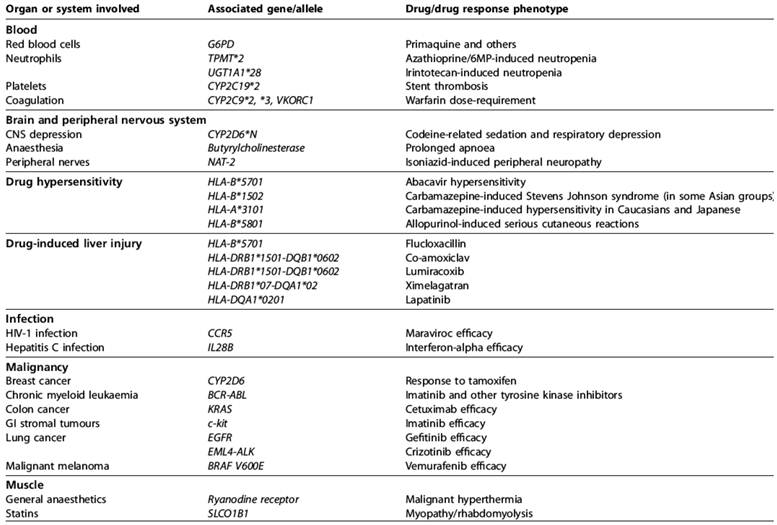
Allele frequencies of variant CYP2D6 alleles (%) in different ethnic populations [209].
| Allele variants | Enzyme function | Caucasian | Asian | Black- African | Ethiopian, Saudi-Arabian |
|---|---|---|---|---|---|
| *1xN, *2xN | gene duplication: increased enzyme activity | 1-5 | 0-2 | 2 | 10-29 |
| *4 | splicing defect: inactive enzyme | 12-21 | 1 | 2 | 1-4 |
| *5 | deletion: no enzyme | 2-7 | 6 | 4 | 1-3 |
| *10 | instable enzyme | 1-2 | 41-51 | 6 | 3-9 |
| *17 | reduced affinity to substrate | 0 | 0 | 20-35 | 3-9 |
| *41 | low protein expression, impaired function | 8.4 | 2.6 |
Most diseases are heterogeneous and the use of molecular diagnostics can divide them into biological subgroups each with their targets and drugs [207].
Consensus on prediction
Our position, and apparently the position of scientists calling for standardization of animal protocols and SRs, that animal models do not currently qualify as predictive modalities for human response to drugs and disease is supported by experts in various fields of science. For example, Alan Oliff, then-executive director for cancer research at Merck Research Laboratories stated: “The fundamental problem in drug discovery for cancer is that the model systems are not predictive at all” [210]. An editorial in Nature Reviews Drug Discovery states: “Clearly, one part of the problem [of drug research] is poorly predictive animal models . . .” [211]. Ellis and Fidler echo this staing: “Preclinical models, unfortunately, seldom reflect the disease state within humans” [212]. Horrobin addressed the use of animal models stating: “Does the use of animal models of disease take us any closer to understanding human disease? With rare exceptions, the answer to this question is likely to be negative” [98]. Fliri pointed out that: “Currently, no method exists for forecasting broad biological activity profiles of medicinal agents even within narrow boundaries of structurally similar molecules” [213]. Speaking of toxicity trials for new drugs in humans, an unnamed clinician was quoted in Science as stating: “If you were to look in [a big company's] files for testing small-molecule drugs you'd find hundreds of deaths” [214]. Frances Collins, director of NIH, has also spoken out on the poor predictive value of animal models [215, 216].
Neuzil et al state: “Animal testing is not ideal either, as the predictive value of such tests is limited owing to metabolic differences between humans and animals, and many ethical issues are raised by the testing” [217]. Cook et al state:
Over many years now there has been a poor correlation between preclinical therapeutic findings and the eventual efficacy of these [anti-cancer] compounds in clinical trials [218, 219]. . . . The development of antineoplastics is a large investment by the private and public sectors, however, the limited availability of predictive preclinical systems obscures our ability to select the therapeutics that might succeed or fail during clinical investigation. [220]
Seidle [221] reported on the conclusions of a conference of experts in toxicology from pharmaceutical companies, contract research companies and others. The consensus was that: “the information obtained from conventional acute toxicity studies is of little or no value in the pharmaceutical development process” [222]. This statement was “subsequently considered and endorsed by regulators and scientists from the EU, US and Japan at a workshop in November 2006 [222].” A survey at the conference [223] revealed that:
- 100% of respondents found data from acute toxicity studies of little or no use and only used the information in dose setting for other studies in exceptional circumstances.
- 100% of respondents agreed that they would not carry out acute toxicity testing if it were not a regulatory requirement.
- 100% of respondents agreed that acute toxicity studies were not used to identify target organs.
- 100% of respondents never use acute toxicity data to help set the starting dose in man.
- 81% of respondents thought the data obtained from acute toxicity studies was of no use to regulators or clinicians. [221]
Sharp and Langer summarized the current situation: “The next challenge for biomedical research will be to solve problems of highly complex and integrated biological systems within the human body. Predictive models of these systems in either normal or disease states are beyond the capability of current knowledge and technology” [224].
We note that the above scientists have not, to the best of our knowledge, agreed with us that animal models are incapable of being predictive modalities. We again attribute this to the fact that the discussion regarding evolved complex systems is relatively new. We also again note that SRs and standardization may contribute to the use of animals in categories 3-9 of table 2. We do not deny that animals can be successfully used for such endeavors in science and research and recognize the value of SRs in improving such uses. However, we have presented a case against expecting animal models to ever be predictive modalities for human response to drugs and disease regardless of improvement in methodology. Even if methodological issues were to prove the problem in some of the studies that reveal PPVs of ~0.5, the lack of studies revealing any animal model to be predictive modality (for example in teratogenicity, carcinogenicity, hepatotoxicity, efficacy for a class of drugs, mechanisms of a class of diseases) is consistent with our theory.
Summary
Animal models have historically been unable to predict human response to drugs and disease and animal-based research has historically displayed methodological problems that make SRs difficult. One proposed solution that would address both problems is standardization of protocols thus permitting SRs of animal models, which would in turn improve the models thus possibly allowing accurate predictions, via high PPV and NPVs, for human response to drugs and disease. We have argued that even if the methodology for animal models could be standardized and subject to SRs, animal models would still fail to be predictive modalities for human response to drugs and disease because of considerations from complexity theory and evolutionary biology. Put succinctly, humans and animals are complex systems with different evolutionary trajectories.
We also reject the notion that a combination of the results of several studies in a SR or meta-analysis may produce information relevant for judging the safety and efficacy of drugs that is not directly visible in the individual animal studies (such as significant side effects or overall efficacy). The problem is that animal models are not predictive modalities, not that animal models fail to reveal side effects. Many side effects from drugs in development are already observed in animal models but there is no predictive value for humans.
As we discussed, SRs are only useful if there is scientific validity to the assumptions or axioms underlying the research. There is no reason to conduct SRs of homeopathy nor does complexity theory and evolutionary biology offer any reason to expect SRs of animal models to be productive. Regardless of how the problem is approached, animal and humans will always be differently complex. Personalized medicine puts this in perspective.
One reason SRs are necessary is that experts are unreliable for evaluating controversies in their own field. We would extend that concept to include the fact that human nature is also problematic when questioning assumptions is required. Tradition, the status quo, “We always do it that way,” resistance to change both individually and in the form of institutional inertia, all combine to challenge those who ask epistemological questions. Financial interests also complicate the situation. Add to all of this the fact that the axioms underlying such practices are not usually discussed among scientists (being I the realm of philosophy of science) and the result is that challenging the axioms upon which these practices are based becomes almost impossible. Nevertheless it is vital to do so in order for science in general, and medical science in particular, to advance.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Van Buskirk NE. The Review Article in MEDLINE: Ambiguity of Definition and Implications for Online Searchers. Bull. Med. Libr. Assoc. 1984;72(4):349-52
2. Greenhalgh T. How to read a paper: Papers that summarise other papers (systematic reviews and meta-analyses). BMJ. 1997;315:672-5 doi:10.1136/bmj.315.7109.672
3. Shermer M. The Believing Brain: From Ghosts and Gods to Politics and Conspiracies---How We Construct Beliefs and Reinforce Them as Truths. New York: Times Books. 2011
4. Green S, Higgins J, Alderson P, Clarke M, Mulrow C, Oxman A. Chapter 1: Introduction. In: (ed.) Higgins J, Green S. Cochrane Handbook for Systematic Reviews of Interventions Version 501. London: The Cochrane Collaboration. 2008
5. The Cochrane Collaboration. Cochrane Handbook for Systematic Reviews of Interventions. London: The Cochrane Collaboration. 2011
6. Sackett DL, Rosenberg WM, Gray JA, Haynes RB, Richardson WS. Evidence based medicine: what it is and what it isn't. BMJ. 1996;312:71-2
7. Glass G. Primary, secondary and meta-analysis of research. Educational Researcher. 1976;5:3-8
8. Barrett S. Be Wary of Acupuncture, Qigong, and "Chinese Medicine". Quackwatch. 2011
9. Novella S. Acupuncture and Acoustic Waves. Neurologica. 2011
10. Gorski D. NIH Director Francis Collins doesn't understand the problem with CAM. Science-Based Medicine. 2012
11. Barret S. Quackwatch; Homeopathy: The Ultimate Fake. Quackwatch. 2009
12. No author listed. Human albumin administration in critically ill patients: systematic review of randomised controlled trials. Cochrane Injuries Group Albumin Reviewers. BMJ. 1998;317:235-40
13. Finfer S, Bellomo R, Boyce N, French J, Myburgh J, Norton R. A comparison of albumin and saline for fluid resuscitation in the intensive care unit. N Engl J Med. 2004;350:2247-56
14. Hooijmans CR, Leenaars M, Ritskes-Hoitinga M. A gold standard publication checklist to improve the quality of animal studies, to fully integrate the Three Rs, and to make systematic reviews more feasible. Alternatives to laboratory animals: ATLA. 2010;38:167-82
15. Nuffield Council on Bioethics. The ethics of research involving animals. London: Nuffield Council on Bioethics. 2005
16. NC3Rs. Systematic Reviews of Animal Research. London: NC3Rs. 2011
17. Macleod MR, O'Collins T, Howells DW, Donnan GA. Pooling of animal experimental data reveals influence of study design and publication bias. Stroke. 2004;35:1203-8
18. Bracken M. Why animal studies are often poor predictors of human reactions to exposure. J R Soc Med. 2008;101:120-22
19. Bracken MB. Why are so many epidemiology associations inflated or wrong? Does poorly conducted animal research suggest implausible hypotheses? Annals of epidemiology. 2009;19:220-4 doi:10.1016/j.annepidem.2008.11.006
20. UMC St Radboud. About systematic reviews. Bereikbaarheid: UMC St Radboud. 2012
21. Kilkenny C, Browne W, Cuthill IC, Emerson M, Altman DG. Animal research: reporting in vivo experiments-The ARRIVE Guidelines. J Cereb Blood Flow Metab. 2011;31:991-3 doi:jcbfm2010220 [pii]10.1038/jcbfm.2010.220
22. Crossley NA, Sena E, Goehler J, Horn J, van der Worp B, Bath PM. et al. Empirical evidence of bias in the design of experimental stroke studies: a metaepidemiologic approach. Stroke. 2008;39:929-34
23. Amarasingh S, Macleod MR, Whittle IR. What is the translational efficacy of chemotherapeutic drug research in neuro-oncology? A systematic review and meta-analysis of the efficacy of BCNU and CCNU in animal models of glioma. Journal of neuro-oncology. 2009;91:117-25 doi:10.1007/s11060-008-9697-z
24. Dirnagl U, Macleod MR. Stroke research at a road block: the streets from adversity should be paved with meta-analysis and good laboratory practice. British Journal of Pharmacology. 2009;157:1154-6
25. Macleod M. Systematic Review and Meta-analysis of Experimental Stroke. International Journal of Neuroprotection and Neuroregeneration. 2004;1:9-12
26. Macleod MR, Ebrahim S, Roberts I. Surveying the literature from animal experiments: systematic review and meta-analysis are important contributions. BMJ. 2005;331:110. doi:331/7508/110-b [pii]10.1136/bmj.331.7508.110-b
27. O'Collins VE, Macleod MR, Cox SF, Van Raay L, Aleksoska E, Donnan GA. et al. Preclinical drug evaluation for combination therapy in acute stroke using systematic review, meta-analysis, and subsequent experimental testing. Journal of cerebral blood flow and metabolism: official journal of the International Society of Cerebral Blood Flow and Metabolism. 2011;31:962-75 doi:10.1038/jcbfm.2010.184
28. Sena E, van der Worp HB, Howells D, Macleod M. How can we improve the pre-clinical development of drugs for stroke? Trends in neurosciences. 2007;30:433-9 doi:10.1016/j.tins.2007.06.009
29. O'Collins VE, Macleod MR, Donnan GA, Horky LL, van der Worp BH, Howells DW. 1,026 experimental treatments in acute stroke. Ann Neurol. 2006;59:467-77 doi:10.1002/ana.20741
30. Frantzias J, Sena ES, Macleod MR, Salman RA-S. Treatment of intracerebral hemorrhage in animal models: Meta-analysis. Annals of Neurology. 2011;69:389-99 doi:10.1002/ana.22243
31. PAO-Heyendael. 1st International Symposium and Workshop on Systematic Reviews in Laboratory Animal Science. PAO-Heyendael. 2012
32. Pound P, Ebrahim S, Sandercock P, Bracken MB, Roberts I. Where is the evidence that animal research benefits humans? BMJ. 2004;328:514-7 doi:10.1136/bmj.328.7438.514 [doi]328/7438/514 [pii]
33. Smith JA, Birke L, Sadler D. Reporting animal use in scientific papers. Laboratory animals. 1997;31:312-7
34. Sniekers YH, Weinans H, Bierma-Zeinstra SM, van Leeuwen JP, van Osch GJ. Animal models for osteoarthritis: the effect of ovariectomy and estrogen treatment - a systematic approach. Osteoarthritis Cartilage. 2008;16:533-41 doi:10.1016/j.joca.2008.01.002
35. Alfaro V. Specification of laboratory animal use in scientific articles: current low detail in the journals' instructions for authors and some proposals. Methods Find Exp Clin Pharmacol. 2005;27:495-502 doi:10.1358/mf.2005.27.7.921309
36. Festing MF. Inbred strains should replace outbred stocks in toxicology, safety testing, and drug development. Toxicologic pathology. 2010;38:681-90 doi:10.1177/0192623310373776
37. Macleod MR, Fisher M, O'Collins V, Sena ES, Dirnagl U, Bath PM. et al. Reprint: Good laboratory practice: preventing introduction of bias at the bench. Journal of cerebral blood flow and metabolism: official journal of the International Society of Cerebral Blood Flow and Metabolism. 2009;29:221-3 doi:10.1038/jcbfm.2008.101
38. van der Worp HB, Howells DW, Sena ES, Porritt MJ, Rewell S, O'Collins V. et al. Can Animal Models of Disease Reliably Inform Human Studies? PLoS Med. 2010;7:e1000245
39. Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 2010;8:e1000412
40. Greek R, Shanks N. FAQs About the Use of Animals in Science: A handbook for the scientifically perplexed. Lanham: University Press of America. 2009
41. Greek R, Greek J. Is the use of sentient animals in basic research justifiable? Philos Ethics Humanit Med. 2010;5:14. doi:1747-5341-5-14 [pii]10.1186/1747-5341-5-14
42. Dombrowski D. Babies and Beasts: The Argument from Marginal Cases: University of Illinois Press. 1997.
43. Dombrowski DA. Is the argument from marginal cases obtuse? J Appl Philos. 2006;23:223-32
44. NABR. The Human care and Treatment of Laboratory Animals. NABR. 1999
45. Council for International Organizations of Medical Sciences (CIMOS). International ethical guidelines for biomedical research involving human subjects. Bull Med Ethics. 2002:17-23
46. Festing S, Wilkinson R. The ethics of animal research. Talking Point on the use of animals in scientific research. EMBO reports. 2007;8:526-30
47. Gad S. Preface. In: Gad S, editor. Animal Models in Toxicology. Boca Rotan: CRC Press. 2007:1-18
48. CNN. Is animal testing necessary? CNN. 2009
49. Marshall BioResources. Benefits of Animal Research. Marshall BioResources. 2010
50. Buzoni-Gatel D, Decelle T, Hardy P, Montagutelli X, Louis J. Animal Models and Relevance/Predictivity: how to better leverage the knowledge of the veterinarian field. Fondation Mérieux. 2011
51. Vassar R. Alzheimer's therapy: a BACE in the hand? Nat Med. 2011;17:932-3
52. Devoy A, Bunton-Stasyshyn RKA, Tybulewicz VLJ, Smith AJH, Fisher EMC. Genomically humanized mice: technologies and promises. Nat Rev Genet. 2012;13:14-20
53. Hau J. Animal Models. In: Hau J, van Hoosier Jr GK, editors. Handbook of Laboratory Animal Science Second Edition Animal Models. 2nd ed. Boca Rotan: CRC Press. 2003:1-9
54. Giles J. Animal experiments under fire for poor design. Nature. 2006;444:981
55. Gibson E. Q&A: Dr. Keith Cheng, researcher at Penn State's College of Medicine, shares views on using animals in scientific research. The Patriot-News. 2012
56. Heywood R. Clinical Toxicity--Could it have been predicted? Post-marketing experience. In: CE Lumley, Walker S, editors. Animal Toxicity Studies: Their Relevance for Man. Lancaster: Quay. 1990:57-67
57. Rudczynski AB. Letter. New Haven Register. New Haven. 2011
58. Sisto A. Italian scientists convicted over earthquake warning. Chicago: Chicago Tribune. 2012
59. Leshner A. Letter to Giorgio Napolitano. AAAS. 2012
60. Hall SS. Scientists on trial: At fault? Nature. 2011;477:264-69 doi:10.1038/477264a
61. Macleod M. Why animal research needs to improve. Nature. 2011;477:511. doi:10.1038/477511a
62. Greek R, Hansen LA, Menache A. An analysis of the Bateson Review of research using nonhuman primates. Medicolegal and Bioethics. 2011;1:3-22
63. Greek R, Menache A, Rice MJ. Animal models in an age of personalized medicine. Personalized Medicine. 2012;9:47-64 doi:10.2217/pme.11.89
64. Greek R, Shanks N, Rice MJ. The History and Implications of Testing Thalidomide on Animals. The Journal of Philosophy, Science & Law. 2011:11
65. Shanks N, Greek R. Animal Models in Light of Evolution. Boca Raton: Brown Walker. 2009
66. Shanks N, Greek R, Greek J. Are animal models predictive for humans? Philos Ethics Humanit Med. 2009;4:2. doi:1747-5341-4-2 [pii]10.1186/1747-5341-4-2
67. Gamble LJ, Matthews QL. Current progress in the development of a prophylactic vaccine for HIV-1. Drug Des Devel Ther. 2010;5:9-26 doi:10.2147/DDDT.S6959
68. Editorial. Cold shower for AIDS vaccines. Nat Med. 2007;13:1389-90
69. van der Worp HB, Macleod MR. Preclinical studies of human disease: Time to take methodological quality seriously. Journal of molecular and cellular cardiology. 2011;51:449-50
70. Dirnagl U, Lauritzen M. Improving the Quality of Biomedical Research: Guidelines for Reporting Experiments Involving Animals. J Cereb Blood Flow Metab. 2011;31:989-90
71. Macleod M, van der Worp HB. Animal models of neurological disease: are there any babies in the bathwater? Practical Neurology. 2010;10:312-4 doi:10.1136/jnnp.2010.230524
72. American Paraplegia Society. Symposium on spinal cord injury models. Presented at the 33rd annual meeting of the American Paraplegia Society. September 1987. J Am Paraplegia Soc. 1988;11:23-58
73. Crowley WF Jr. Translation of basic research into useful treatments: how often does it occur? Am J Med. 2003;114:503-5
74. Enna SJ, Williams M. Defining the role of pharmacology in the emerging world of translational research. Advances in pharmacology. 2009;57:1-30 doi:10.1016/S1054-3589(08)57001-3
75. Hackam DG, Redelmeier DA. Translation of research evidence from animals to humans. JAMA. 2006;296:1731-2
76. Schnabel J. Neuroscience: Standard model. Nature. 2008;454:682-5
77. Hurko O. Understanding the strategic importance of biomarkers for the discovery and early development phases. Drug Discovery World. 2006:63-74
78. Cressey D. Traditional drug-discovery model ripe for reform. Nature. 2011;471:17-8
79. Hughes JP, Rees S, Kalindjian SB, Philpott KL. Principles of early drug discovery. British Journal of Pharmacology. 2011;162:1239-49
80. Giri S, Bader A. Foundation review: Improved preclinical safety assessment using micro-BAL devices: the potential impact on human discovery and drug attrition. Drug Discovery Today. 2011;16:382-97
81. Litchfield JT Jr. Symposium on clinical drug evaluation and human pharmacology. XVI. Evaluation of the safety of new drugs by means of tests in animals. Clin Pharmacol Ther. 1962;3:665-72
82. Suter K. What can be learned from case studies? The company approach. In: Lumley C, Walker S, editors. Animal Toxicity Studies: Their Relevance for Man. Lancaster: Quay. 1990:71-8
83. Lumley C. Clinical toxicity: could it have been predicted? Premarketing experience. In: Lumley C, Walker S, editors. Animal Toxicity Studies: Their Relevance for Man. London: Quay. 1990:49-56
84. Spriet-Pourra C, Auriche M. SCRIP Reports: PJB. 1994.
85. Eason CT, Bonner FW, Parke DV. The importance of pharmacokinetic and receptor studies in drug safety evaluation. Regul Toxicol Pharmacol. 1990;11:288-307
86. Igarashi T. The duration of toxicity studies required to support repeated dosing in clinical investigation—A toxicologists opinion. In: C Parkinson NM, C Lumley, SR Walker, editor. CMR Workshop: The Timing of Toxicological Studies to Support Clinical Trials. Boston/UK: Kluwer. 1994:67-74
87. Igarashi T, Nakane S, Kitagawa T. Predictability of clinical adverse reactions of drugs by general pharmacology studies. J Toxicol Sci. 1995;20:77-92
88. Igarashi T, Yabe T, Noda K. Study design and statistical analysis of toxicokinetics: a report of JPMA investigation of case studies. J Toxicol Sci. 1996;21:497-504
89. Weaver JL, Staten D, Swann J, Armstrong G, Bates M, Hastings KL. Detection of systemic hypersensitivity to drugs using standard guinea pig assays. Toxicology. 2003;193:203-17 doi:S0300483X03002671 [pii]
90. Willis RC. The Virtual Patient. Modern Drug Discovery. 2003;6:35-40
91. Sankar U. The Delicate Toxicity Balance in Drug Discovery. The Scientist. 2005;19:32
92. Fourches D, Barnes JC, Day NC, Bradley P, Reed JZ, Tropsha A. Cheminformatics analysis of assertions mined from literature that describe drug-induced liver injury in different species. Chem Res Toxicol. 2010;23:171-83
93. Park BK, Boobis A, Clarke S, Goldring CEP, Jones D, Kenna JG. et al. Managing the challenge of chemically reactive metabolites in drug development. Nat Rev Drug Discov. 2011;10:292-306
94. Lin JH. Species similarities and differences in pharmacokinetics. Drug Metab Dispos. 1995;23:1008-21
95. Sietsema WK. The absolute oral bioavailability of selected drugs. Int J Clin Pharmacol Ther Toxicol. 1989;27:179-211
96. Greek R. Animal Models and the Development of an HIV Vaccine. J AIDS Clinic Res. 2012 S8:001
97. Greek R. Book Review. Zoobiquity: What Animals Can Teach Us About Health and the Science of Healing. Animals. 2012;2:559-63
98. Horrobin DF. Modern biomedical research: an internally self-consistent universe with little contact with medical reality? Nat Rev Drug Discov. 2003;2:151-4
99. Wall RJ, Shani M. Are animal models as good as we think? Theriogenology. 2008;69:2-9
100. Zielinska E. Building a better mouse. The Scientist. 2010;24:34-8
101. Miska D. Biotech's twentieth birthday blues. Nat Rev Drug Discov. 2003;2:231-3
102. Ahn AC, Tewari M, Poon CS, Phillips RS. The limits of reductionism in medicine: could systems biology offer an alternative? PLoS Med. 2006;3:e208
103. Krakauer DC, Collins JP, Erwin D, Flack JC, Fontana W, Laubichler MD. et al. The challenges and scope of theoretical biology. J Theor Biol. 2011;276:269-76
104. American Association for the Advancement of Science. Q & A on Evolution and Intelligent Design. Washington DC: American Association for the Advancement of Science. 2011
105. Van Regenmortel MHV. Basic Research in HIV vaccinology is hampered by reductionist thinking. Frontiers in Immunology. 2012:3. doi:10.3389/fimmu.2012.00194
106. Jura J, Wegrzyn P, Koj A. Regulatory mechanisms of gene expression: complexity with elements of deterministic chaos. Acta Biochim Pol. 2006;53:1-10 doi:20061177 [pii]
107. Vicsek T. The bigger picture. Nature. 2002;418:131
108. Goodman AF, Bellato CM, Khidr L. The Uncertain Future for Central Dogma. Uncertainty serves as a bridge from determinism and reductionism to a new picture of biology. The Scientist. 2005:19
109. Van Regenmortel M. Reductionism and complexity in molecular biology. Scientists now have the tools to unravel biological complexity and overcome the limitations of reductionism. EMBO Rep. 2004;5:1016-20
110. Csete ME, Doyle JC. Reverse engineering of biological complexity. Science. 2002;295:1664-9
111. Sole R, Goodwin B. Signs of Life: How Complexity Pervades Biology: Basic Books. 2002.
112. Kitano H. Computational systems biology. Nature. 2002;420:206-10
113. Kitano H. Systems biology: a brief overview. Science. 2002;295:1662-4
114. Kauffman SA. The Origins of Order: Self-Organization and Selection in Evolution. Oxford University Press. 1993
115. Ottino JM. Engineering complex systems. Nature. 2004;427:399
116. Alm E, Arkin AP. Biological networks. Curr Opin Struct Biol. 2003;13:193-202
117. Goodwin B. How the Leopard Changed Its Spots: The Evolution of Complexity. Princeton: Princeton University Press. 2001
118. Van Regenmortel M. Reductionism and the search for structure-function relationships in antibody molecules. J Mol Recognit. 2002;15:240-7 doi:10.1002/jmr.584
119. van Regenmortel M. Pitfalls of Reductionism in Immunology. In: van Regenmortel M, Hull DL, editors. Promises and Limits of Reductionism in the Biomedical Sciences. Chichester: John Wiley & Sons LTD. 2002:47-66
120. van Regenmortel M. Biological complexity emerges from the ashes of genetic reductionism. Journal of Molecular Recognition. 2004;17:145-8
121. Cairns-Smith AG. Seven Clues to the Origin of Life: A Scientific Detective Story. Cambridge: Cambridge University Press. 1986
122. Monte J, Liu M, Sheya A, Kitami T. Definitions, Measures, and Models of Robustness in Gene Regulatory Network. Report of research work for CSSS05. 2005
123. Morowitz HJ. The Emergence of Everything: How the World Became Complex. Oxford: Oxford University Press. 2002
124. Novikoff AB. The Concept of Integrative Levels and Biology. Science. 1945;101:209-15
125. Van Regenmortel MH, Hull DL. Promises and Limits of Reductionism in the Biomedical Sciences (Catalysts for Fine Chemical Synthesis). West Sussex: Wiley. 2002
126. Woodger JH. Biological Principles. New York: Humanities Press. 1967
127. Kastens KA, Manduca CA, Cervato C, Frodeman R, Goodwin C, Liben LS. et al. How Geoscientists Think and Learn. Eos Trans AGU. 2009:90. doi:10.1029/2009eo310001
128. Greek R, Rice MJ. Animal models and conserved processes. Theoretical Biology and Medical Modelling. 2012:9. doi:10.1186/1742-4682-9-40
129. LaFollette H, Shanks N. Animal models in biomedical research: some epistemological worries. Public Aff Q. 1993;7:113-30
130. LaFollette H, Shanks N. Animal Experimentation: The Legacy of Claude Bernard. International Studies in the Philosophy of Science. 1994;8:195-210
131. LaFollette H, Shanks N. Two Models of Models in Biomedical Research. Philosophical Quarterly. 1995;45:141-60
132. Mazzocchi F. Complexity and the reductionism-holism debate in systems biology. Wiley Interdiscip Rev Syst Biol Med. 2012;4:413-27 doi:10.1002/wsbm.1181
133. Morange M. A successful form for reductionism. The Biochemist. 2001;23:37-9
134. Pearson H. Surviving a knockout blow. Nature. 2002;415:8-9 doi:10.1038/415008a415008a [pii]
135. Willyard C. HIV gender clues emerge. Nat Med. 2009;15:830. doi:nm0809-830b [pii]10.1038/nm0809-830b
136. Holden C. Sex and the suffering brain. Science. 2005;308:1574. doi:308/5728/1574 [pii]10.1126/science.308.5728.1574
137. Kaiser J. Gender in the pharmacy: does it matter? Science. 2005;308:1572. doi:308/5728/1572 [pii]10.1126/science.308.5728.1572
138. Simon V. Wanted: women in clinical trials. Science. 2005;308:1517. doi:308/5728/1517 [pii]10.1126/science.1115616
139. Wald C, Wu C. Of Mice and Women: The Bias in Animal Models. Science. 2010;327:1571-2
140. Klein S, Huber S. Sex differences in susceptibility to viral infection. In: Klein S, Roberts C, editors. Sex hormones and immunity to infection. Berlin: Springer-Verlag. 2010:93-122
141. Cheung DS, Warman ML, Mulliken JB. Hemangioma in twins. Ann Plast Surg. 1997;38:269-74
142. Gregor Z, Joffe L. Senile macular changes in the black African. Br J Ophthalmol. 1978;62:547-50
143. Stamer UM, Stuber F. The pharmacogenetics of analgesia. Expert Opin Pharmacother. 2007;8:2235-45 doi:10.1517/14656566.8.14.2235
144. Wilke RA, Dolan ME. Genetics and Variable Drug Response. JAMA: The Journal of the American Medical Association. 2011;306:306-7 doi:10.1001/jama.2011.998
145. Kopp JB, Nelson GW, Sampath K, Johnson RC, Genovese G, An P. et al. APOL1 Genetic Variants in Focal Segmental Glomerulosclerosis and HIV-Associated Nephropathy. Journal of the American Society of Nephrology. 2011;22:2129-37 doi:10.1681/asn.2011040388
146. Haiman CA, Stram DO, Wilkens LR, Pike MC, Kolonel LN, Henderson BE. et al. Ethnic and racial differences in the smoking-related risk of lung cancer. N Engl J Med. 2006;354:333-42 doi:354/4/333 [pii]10.1056/NEJMoa033250
147. Spielman RS, Bastone LA, Burdick JT, Morley M, Ewens WJ, Cheung VG. Common genetic variants account for differences in gene expression among ethnic groups. Nat Genet. 2007;39:226-31 doi:ng1955 [pii]10.1038/ng1955
148. Couzin J. Cancer research. Probing the roots of race and cancer. Science. 2007;315:592-4 doi:315/5812/592 [pii]10.1126/science.315.5812.592
149. Kalow W. Interethnic variation of drug metabolism. Trends in Pharmacological Sciences. 1991;12:102-7
150. Bruder CE, Piotrowski A, Gijsbers AA, Andersson R, Erickson S, de Stahl TD. et al. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am J Hum Genet. 2008;82:763-71 doi:S0002-9297(08)00102-X [pii]10.1016/j.ajhg.2007.12.011
151. Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J. et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Research. 2010;20:170-9 doi:10.1101/gr.100289.109
152. Wong AH, Gottesman II, Petronis A. Phenotypic differences in genetically identical organisms: the epigenetic perspective. Hum Mol Genet. 2005;1:R11-8 doi:14/suppl_1/R11 [pii]10.1093/hmg/ddi116
153. Dempster EL, Pidsley R, Schalkwyk LC, Owens S, Georgiades A, Kane F. et al. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Human molecular genetics. 2011;20:4786-96 doi:10.1093/hmg/ddr416
154. Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML. et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604-9 doi:0500398102 [pii]10.1073/pnas.0500398102
155. Williams G. Chaos Theory Tamed. Washington, DC: Joseph Henry Press. 1997
156. Bickel PJ, Buhlmann P. What is a linear process? Proceedings of the National Academy of Sciences of the United States of America. 1996;93:12128-31
157. Lippman N, Stein KM, Lerman BB. Nonlinear forecasting and the dynamics of cardiac rhythm. J Electrocardiol. 1995;28(Suppl):65-70
158. Rosen R. Essays on Life Itself. New York: Columbia University Press. 1998
159. Koch C. Systems biology. Modular biological complexity. Science. 2012;337:531-2
160. Pelak K, Need AC, Fellay J, Shianna KV, Feng S, Urban TJ. et al. Copy number variation of KIR genes influences HIV-1 control. PLoS biology. 2011;9:e1001208
161. Mittelstrass K, Ried JS, Yu Z, Krumsiek J, Gieger C, Prehn C. et al. Discovery of sexual dimorphisms in metabolic and genetic biomarkers. PLoS genetics. 2011;7:e1002215
162. Yucesoy B, Johnson VJ, Fluharty K, Kashon ML, Slaven JE, Wilson NW. et al. Influence of cytokine gene variations on immunization to childhood vaccines. Vaccine. 2009;27:6991-7
163. King C. Personalised vaccines could protect all children. New Scientist. 2009:11
164. Sivakumaran S, Agakov F, Theodoratou E, Prendergast JG, Zgaga L, Manolio T. et al. Abundant pleiotropy in human complex diseases and traits. American journal of human genetics. 2011;89:607-18
165. Gustincich S, Sandelin A, Plessy C, Katayama S, Simone R, Lazarevic D. et al. The complexity of the mammalian transcriptome. J Physiol. 2006;575:321-32
166. Burgess DJ. Evo-devo: Hidden rewiring comes to light. Nat Rev Genet. 2011;12:586-7
167. Lowe CB, Kellis M, Siepel A, Raney BJ, Clamp M, Salama SR. et al. Three periods of regulatory innovation during vertebrate evolution. Science. 2011;333:1019-24
168. Pai AA, Bell JT, Marioni JC, Pritchard JK, Gilad Y. A Genome-Wide Study of DNA Methylation Patterns and Gene Expression Levels in Multiple Human and Chimpanzee Tissues. PLoS Genet. 2011;7:e1001316
169. Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS. et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743-7
170. Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA. et al. Genetic structure of human populations. Science. 2002;298:2381-5
171. Storey JD, Madeoy J, Strout JL, Wurfel M, Ronald J, Akey JM. Gene-expression variation within and among human populations. Am J Hum Genet. 2007;80:502-9
172. Zhang W, Duan S, Kistner EO, Bleibel WK, Huang RS, Clark TA. et al. Evaluation of genetic variation contributing to differences in gene expression between populations. Am J Hum Genet. 2008;82:631-40
173. Pritchard C, Coil D, Hawley S, Hsu L, Nelson PS. The contributions of normal variation and genetic background to mammalian gene expression. Genome Biol. 2006;7:R26
174. Rifkin SA, Kim J, White KP. Evolution of gene expression in the Drosophila melanogaster subgroup. Nat Genet. 2003;33:138-44
175. Sandberg R, Yasuda R, Pankratz DG, Carter TA, Del Rio JA, Wodicka L. et al. Regional and strain-specific gene expression mapping in the adult mouse brain. Proc Natl Acad Sci U S A. 2000;97:11038-43
176. Suzuki Y, Nakayama M. Differential profiles of genes expressed in neonatal brain of 129X1/SvJ and C57BL/6J mice: A database to aid in analyzing DNA microarrays using nonisogenic gene-targeted mice. DNA Res. 2003;10:263-75
177. Somel M, Liu X, Tang L, Yan Z, Hu H, Guo S. et al. MicroRNA-Driven Developmental Remodeling in the Brain Distinguishes Humans from Other Primates. PLoS Biol. 2011;9:e1001214
178. Puente XS, Velasco G, Gutierrez-Fernandez A, Bertranpetit J, King MC, Lopez-Otin C. Comparative analysis of cancer genes in the human and chimpanzee genomes. BMC Genomics. 2006;7:15
179. Varki NM, Strobert E, Dick EJ, Benirschke K, Varki A. Biomedical Differences Between Human and Nonhuman Hominids: Potential Roles for Uniquely Human Aspects of Sialic Acid Biology. Annual Review of Pathology: Mechanisms of Disease. 2011;6:365-93
180. Walker CM. Comparative features of hepatitis C virus infection in humans and chimpanzees. Springer Semin Immunopathol. 1997;19:85-98
181. Gagneux P, Muchmore EA. The chimpanzee model: contributions and considerations for studies of hepatitis B virus. Methods in molecular medicine. 2004;96:289-318
182. Bettauer RH. Chimpanzees in hepatitis C virus research: 1998-2007. Journal of Medical Primatology. 2010;39:9-23
183. Caldwell J. Problems and opportunities in toxicity testing arising from species differences in xenobiotic metabolism. Toxicology letters. 1992:64-65
184. Festing MF. Rat Genetics and Toxicology. In: National Research Council, editor. Microbial Status and Genetic Evaluation of Mice and Rats: Proceedings of the 1999 US/Japan Conference: ILAR. 2000:97
185. Roses AD. Pharmacogenetics and the practice of medicine. Nature. 2000;405:857-65 doi:10.1038/35015728
186. Willyard C. Blue's clues. Nat Med. 2007;13:1272-3
187. Weinshilboum R. Inheritance and drug response. N Engl J Med. 2003;348:529-37
188. Bates S. Progress towards personalized medicine. Drug Discovery Today. 2010;15:115-20
189. Bhathena A, Spear BB. Pharmacogenetics: improving drug and dose selection. Curr Opin Pharmacol. 2008;8:639-46
190. Blair E. Predictive tests and personalised medicine. Drug Discovery World. 2009:27-31
191. Dolgin E. Big pharma moves from 'blockbusters' to 'niche busters'. Nat Med. 2010;16:837
192. Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA. et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809-19 doi:10.1056/NEJMoa1002011
193. Froehlich TE, Epstein JN, Nick TG, Melguizo Castro MS, Stein MA, Brinkman WB. et al. Pharmacogenetic Predictors of Methylphenidate Dose-Response in Attention-Deficit/Hyperactivity Disorder. Journal of the American Academy of Child and Adolescent Psychiatry. 2011;50:1129-39.e2
194. Hudson KL. Genomics, Health Care, and Society. New England Journal of Medicine. 2011;365:1033-41
195. Hughes AR, Spreen WR, Mosteller M, Warren LL, Lai EH, Brothers CH. et al. Pharmacogenetics of hypersensitivity to abacavir: from PGx hypothesis to confirmation to clinical utility. Pharmacogenomics J. 2008;8:365-74 doi:tpj20083 [pii]10.1038/tpj.2008.3
196. Serrano D, Lazzeroni M, Zambon CF, Macis D, Maisonneuve P, Johansson H. et al. Efficacy of tamoxifen based on cytochrome P450 2D6, CYP2C19 and SULT1A1 genotype in the Italian Tamoxifen Prevention Trial. Pharmacogenomics J. 2011;11:100-7
197. Wang D, Guo Y, Wrighton SA, Cooke GE, Sadee W. Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharmacogenomics J. 2011;11:274-86
198. Canto JG, Rogers WJ, Goldberg RJ, Peterson ED, Wenger NK, Vaccarino V. et al. Association of Age and Sex With Myocardial Infarction Symptom Presentation and In-Hospital Mortality. JAMA: The Journal of the American Medical Association. 2012;307:813-22
199. Darlison MG, Pahal I, Thode C. Consequences of the evolution of the GABA(A) receptor gene family. Cell Mol Neurobiol. 2005;25:607-24 doi:10.1007/s10571-005-4004-4
200. Geerts H. Of mice and men: bridging the translational disconnect in CNS drug discovery. CNS Drugs. 2009;23:915-26
201. Jankovic J, Noebels JL. Genetic mouse models of essential tremor: are they essential? J Clin Invest. 2005;115:584-6
202. Kieburtz K, Olanow CW. Translational experimental therapeutics: The translation of laboratory-based discovery into disease-related therapy. Mt Sinai J Med. 2007;74:7-14
203. Liu Z, Maas K, Aune TM. Comparison of differentially expressed genes in T lymphocytes between human autoimmune disease and murine models of autoimmune disease. Clin Immunol. 2004;112:225-30
204. Miklos GLG. The human cancer genome project--one more misstep in the war on cancer. Nat Biotechnol. 2005;23:535-7
205. PMC. Personalized Medicine. Personalized Medicine Coalition. 2006
206. Personalized Medicine Coalition. The Case for Personalized Medicine. Personalized Medicine Coalition. 2011
207. Jørgensen JT. A challenging drug development process in the era of personalized medicine. Drug Discovery Today. 2011;16:891-7
208. Pirmohamed M. Pharmacogenetics: past, present and future. Drug Discovery Today. 2011;16:852-61
209. Stamer UM, Stuber F. Genetic factors in pain and its treatment. Current opinion in anaesthesiology. 2007;20:478-84 doi:10.1097/ACO.0b013e3282ef6b2c
210. Gura T. Cancer Models: Systems for identifying new drugs are often faulty. Science. 1997;278:1041-2
211. Editorial. The time is now. Nat Rev Drug Discov. 2005;4:613
212. Ellis LM, Fidler IJ. Finding the tumor copycat. Therapy fails, patients don't. Nat Med. 2010;16:974-5 doi:nm0910-974 [pii]10.1038/nm0910-974
213. Fliri AF, Loging WT, Thadeio PF, Volkmann RA. Biological spectra analysis: Linking biological activity profiles to molecular structure. Proc Natl Acad Sci U S A. 2005;102:261-6
214. Marshall E. Gene therapy on trial. Science. 2000;288:951-7
215. Collins FS. Reengineering Translational Science: The Time Is Right. Science Translational Medicine. 2011;3:90cm17
216. Reuters. US to develop chip that tests if a drug is toxic. Reuters. 2011
217. Neuzil P, Giselbrecht S, Lange K, Huang TJ, Manz A. Revisiting lab-on-a-chip technology for drug discovery. Nature Reviews Drug Discovery. 2012;11:620-32
218. Johnson JI, Decker S, Zaharevitz D, Rubinstein LV, Venditti JM, Schepartz S. et al. Relationships between drug activity in NCI preclinical in vitro and in vivo models and early clinical trials. Br J Cancer. 2001;84:1424-31
219. Suggitt M, Bibby MC. 50 years of preclinical anticancer drug screening: empirical to target-driven approaches. Clinical cancer research: an official journal of the American Association for Cancer Research. 2005;11:971-81
220. Cook N, Jodrell DI, Tuveson DA. Predictive in vivo animal models and translation to clinical trials. Drug Discovery Today. 2012;17:253-60
221. Seidle T. Opportunities and Barriers to the Replacement of Animals in Acute Systemic Toxicity Testing. AltTox.org. 2007
222. NC3Rs. News: Challenging the requirement for acute toxicity studies - workshop report published. 2007.
223. Chapman K, Robinson S. Challenging the requirement for acute toxicity studies in the development of new medicines. London: NC3Rs. 2007
224. Sharp PA, Langer R. Promoting Convergence in Biomedical Science. Science. 2011;333:527
Author contact
![]() Corresponding author: Ray Greek, Americans For Medical Advancement, 2251 Refugio Rd, Goleta, CA 93117, 805-685-6812. DrRayGreekcom.
Corresponding author: Ray Greek, Americans For Medical Advancement, 2251 Refugio Rd, Goleta, CA 93117, 805-685-6812. DrRayGreekcom.

 Global reach, higher impact
Global reach, higher impact