Impact Factor
ISSN: 1449-1907
Int J Med Sci 2009; 6(6):365-373. doi:10.7150/ijms.6.365 This issue Cite
Research Paper
Autofluorescent Proteins as Photosensitizer in Eukaryontes
1. German Cancer Research Center, Dept. of Biophysics of Macromolecules, INF 580, D-69120 Heidelberg, Germany
2. German Cancer Research Center, Dept. of Medical Physics in Radiology, INF 280, D-69120 Heidelberg, Germany
3. German Cancer Research Center, Core Facility Light Microscopy, INF 581, D-69120 Heidelberg, Germany
Received 2009-9-10; Accepted 2009-11-25; Published 2009-12-1
Abstract
Since the discovery of the green fluorescent green protein (GFP) in 1961 many variants of fluorescent proteins (FP) were detected. The importance was underlined by the Nobel price award in chemistry 2008 for the invention, application, and development of the GFP by Shimomura, Chalfie and Tsien. GFP, first described by Shimomura now is indispensible in the scientific daily life.
Since then and also in future fluorescent proteins will lead to new applications as reporters in cell biology. Such FPs can absorb visible day-light and predominantly one variant of the red fluorescent protein, the KillerRed protein (KRED) emits active electrons producing reactive oxygen species (ROS) leading to photokilling processes in eukaryotes. KRED can be activated by daylight as a photosensitizing agent. It is quite obvious that the KRED's expression and localization is critical with respect to damage, mutation and finally killing of eukaryotic cells. We found evidence that the KRED's cytotoxicity is ascendantly location-dependent from the cell membrane over the nuclear lamina to the chromatin in the cell nucleus. Daylight illumination of cells harbouring the KRED protein fused with the histone H2A, a DNA-binding protein which is critical for the formation of the chromatin structure results in cell killing. Therefore the H2A-KRED fusion protein can be considered as an appropriate candidate for the photodynamic therapy (PDT). This finding can be transferred to current photodynamic approaches and can enhance their therapeutic outcome.
Keywords: Melanoma, fluorescent Proteins, KillerRed, Photo-Dynamic-Therapy (PDT), ROS, Skin Tumors, subcellular Localization, topical Application
Introduction
Without doubt oxygen is considered as a pivotal element for the existence of aerobic life on earth. But in the last forty years, evidences indicated increasingly Janus-faced behaviors of this element1-3 for the following reasons: Under certain conditions, oxygen may produce reactive species, even free radicals responsible for different molecular cell response like cellular stress4,5. Despite all undesired consequences provoked by these oxygen's properties, these facts were not yet in the focus of the scientific discussion and still poorly understood during the last few years as illustrated comprehensively6. The paradox of the oxygen atom depending on its peculiar electronic structure is the existence as a free radical, because the outer valence shell contains one unpaired electron. After combining two oxygen atoms to form molecular oxygen no formation of a spin-pair is possible and resulting in a formation of a bi-radical which allows a stepwise one electron reduction as depicted in Figure 1. Up to this point it's just a non-enzymatic pathway of oxygen reduction results in the generation of different highly reactive intermediates referred as reactive oxygen species (ROS).
Additionally, reactive nitrogen species, such as nitric oxide and peroxynitrite, are biologically relevant O2 derivatives increasingly being recognized as important in vascular biology potential7. Starting with O2 the first one-electron reduction leads to the superoxide anion radical formation (•O2-). After addition of an electron and two protons the highly active species hydrogen peroxide is built. The addition of a further electron results in the hydroxyl radical formation simultaneously releasing a hydroxide anion. The fourth electron addition produces a water molecule. This indicates the role of oxygen as a basis in collecting electrons8.
The figure exemplifies the generation of ROS and their influence on downstream targets in vascular cells. ROS influence a multifaced range of cellular activities e.g. of protein tyrosine phosphatases (PTP). ROS influence gene and protein expression by activating transcription factors, such as NFκB and AP-1. ROS stimulate Ca2+ channels leading to increased [Ca2+]. ROS influence matrix metalloproteinases (MMPs), modulating extracellular matrix protein (ECM) degradation. (Modified according to Touyz and Filomeni) [Touyz, R. M. Antioxid. Redox. Signal. 2005, 7 (9-10), 1302-1314; Filomeni, G.; et al. Biochem. Pharmacol. 2002, 64 (5-6), 1057-1064].

A look behind the origination of aerobic life and the impact of the oxygen could contribute to a better understanding of the oxygen paradox. Despite all barren and hostile circumstances, the aerobic life on earth began under simultaneous evolution of efficient anti ROS-weapon systems like antioxidants and scavengers by which all creatures are extensively endowed. The intracellular redox state is determined by the contribution of different redox couples, at which each couple can exchange electrons in such a way that, by giving or accepting reducing equivalents, may represent cofactors in redox enzymatic reactions. Furthermore, the activator protein 1 (AP-1) the nuclear factor-κB (NF-κB) and protein tyrosine phosphatases (PTPs) are considered as excellent examples, as illustrated in Figure 1 9. The consequences of oxygen activation in human bodies are indeed increasingly observed but only partly recognized, in spite of extensive scientific research on theoretical, experimental and clinical domains10.
In contrast to the prokaryotes the impact of "reactive oxygen species" on the behavior of eukaryotes seems to be better investigated, as shown by searching on the NCBI database PubMed from 07 10 2009. Using the search terms like “eukaryotes” and "reactive oxygen species" cited 1993 for the first time until today, 142 hits were found which it's not very extensive. Especially the fact that 44 articles thereof were published in the last two years suggests an increased scientific interest on pharmacologically inactive molecules which are converted after activation by daylight to photosensitizing agents and which are able to damage, mutate and finally kill eukaryotic cells.
It is documented that fluorescent compounds which absorb daylight around 500 to 700 nm can emit active electrons producing ROS11 which in turn induce cell killing of prokaryotic cells.
Fluorescent proteins (FPs)12 stand for a group of ROS producers. They are originally represented by green fluorescent protein (EGFP)12 which is considered as a promising source of excellent tools for successful live-cell imaging13. Indeed hampering features like quenching effects which can change the EGFP's fluorescent properties were observed14,15. Additionally an oxidant-induced cell death in yeast and in saccharomyces cerevisiae is documented16,17. In comparison, in case of radical or reactive oxygen formation, the amount of data investigations about prokaryotes and eucaryotes expressing FPs is still moderate. Further investigations of the impact of ROS on the acceleration of cellular aging initiated by cellular stress should be extendet to healthy and neoplastic eukaryotes. To investigate ROS influence on cells, we expressed the fluorescent KillerRed (KRED) protein18 from stably transfected plasmids (described in the methods part) in the human HeLa cervix carcinoma, and the human DU145 prostate cancer cell lines.
Our plasmids coding for this red protein contain the sequence of the hydrozoan chromoprotein anm2CP gene (GenBank, accession number AY485336) originating from an Anthomedusa, which transcribes and translates the KillerRed protein (KRED)19. As shown in the literature this KillerRed protein is supposed to produce enough ROS to kill half of the transfected human kidney cells, after 10 minutes illumination. Localizing the KRED protein to mitochondria resulted in an increased cytotoxic efficiency with the greatest extent after 45 minutes20.
This KRED protein, comprehensively described by the Bulina and Lukyanov groups, is exemplified as the first genetically encoded photosensitizer18.
Our intention was to find out whether FPs with different light absorption properties reveal the same ROS producing capacity and looked for a dependency of their intracellular localizations.
Therefore we carried out cell death studies by FP-imaging where the reporter proteins were placed on different intracellular structural locations. We observed in HeLa cervix carcinoma and DU 145 human prostate cancer cells stably expressing KRED daylight-induced cell toxicity with the confocal laser scanning microscopy (CLSM).
Cell toxic effects caused by KRED after white light exposition are already documented in eukaryotes20 but data concerning the different damaging sensitivity to visible light depending on the location of the reporter remain to be answered. Our data indicate different FP's toxicity, depending on its intracellular localization.
Material & Methods
Plasmid vectors constructions
For the investigation of the subcellular localization dependant cell toxicity FP-induced we used the following purchasable and recombined fusion-vectors. As a reference the pEGFP vector was used (Figure 2).
a) pEGFP vector
The figure displays the physical map of the pEGFP a red-shifted variant of the WT GFP optimized for higher fluorescence and higher expression in mammalian cells. GenBank Acc. No.: #U55762. (Details see Clontech user manual http://www.clontech.com/images/pt/dis_vectors/PT3027-5.pdf)
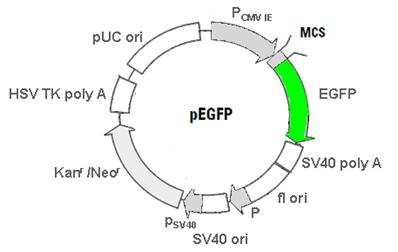
b) pKillerRed vector - Free KRED protein
This physical map shows the body of the pKillerRed mammalian expression vector encoding the red fluorescent protein KillerRed alone in eukaryotic (mammalian) cells (Evrogen FP961; GenBank Acc. No.: AY969116). (Details see Evrogen user manual http://www.evrogen.com/products/KillerRed/KillerRed_Related_products.shtml)
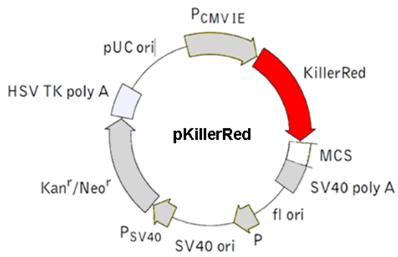
c) pKillerRed-mem vector - Membrane-located KRED protein
The figure illustrates the pKillerRed-mem a mammalian expression vector which encodes membrane-targeted KillerRed (Cat. No.: #FP966). (Details see Evrogen user manual http://www.evrogen.com/products/KillerRed/KillerRed_Related_products.shtml)  shows the mem sequence.
shows the mem sequence.
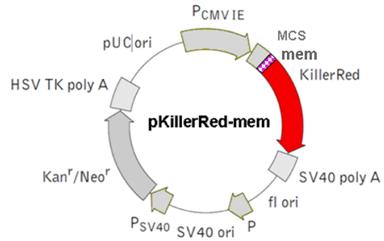
d) pKillerRed Lamin B1 vector - Lamin B1-localized KRED protein
Physical map of the vector expressing the fusion protein KRED-Lamin B1.  The Lamin B1 was inserted into the MCS. (The Lamin B1 sequence was kindly provided by Harald Herrmann, this institute)
The Lamin B1 was inserted into the MCS. (The Lamin B1 sequence was kindly provided by Harald Herrmann, this institute)
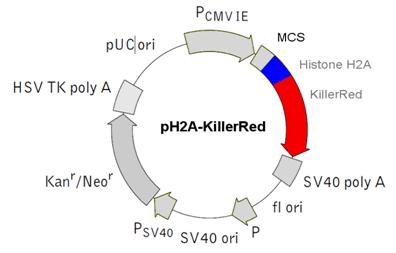
e) pH2A Histone-KillerRed vector - Histone H2A-localized KRED protein
Physical map of the vector expressing the histone fusion protein H2A-KRED.  The histone H2A was inserted into the MCS.
The histone H2A was inserted into the MCS.
Transfection
HeLa cervix carcinoma and DU 145 human prostate cancer cells were transfected with plasmids expressing differently coloured autofluorescent proteins according to the Fugen HD's user manual (Roche, Germany). Stable transformations with the mentioned plasmid constructs were generated over weeks by selection pressure in cell culture with 500 µg/ml G-418 (Geneticin) final concentration. Clones were picked, cultured and used in the experiments.
Cell culture
Cell clones were cultured and maintained in RPMI medium (Gibco, USA) supplemented with FCS 10% (Biochrom, Germany) and L-glutamin 200 mM (Biochrom, Germany) at 37°C in a humid 5% CO2 atmosphere. The cultures were visibly green, yellow or red. Near confluency, the cells were washed with HBSS (Hank's balanced salt concentration, PAN, Germany). After trypsinization (0.5%) the cells were harvested in RPMI with 2% FCS and centrifuged (800 U/ min, 5 minutes; Hereaus, Germany). After resuspension of the cell pellet in HBSS the cell number was adjusted with HBSS to 1 × 106 cells × ml-1 for further experiments.
Illumination of the KRED expressing cells
HeLa and DU 145 cells, grown in RPMI-medium were transferred to quartz cuvettes (HELLMA, Germany). These cuvettes were placed under one full-spectrum sunlight bulb with 32 Watt (www.androv-medical.de) in a distance of 1 cm.
The illumination took place at room temperature; the quartz cuvettes were placed on an aluminium block, cooled by a fan to keep the room temperature. The 32 Watt bulb has a measured intensity of 20.000 lux in a 1cm distance, which reflects a normal daylight in a cloudy summer21. We used the following time points: 15, 30, 60, 90, 120, and 180 minutes for the measurements of the clones. Transfected cells were examined under identical conditions, controls were measured without illumination.
Subcellular localization of the FPs by confocal laser scanning microscopy (CLSM)
To perform confocal laser scanning microscopic (CLSM) studies, DU 145 and HeLa cells (2 × 104) were seeded into chambered cover class (Nunc 8-Well, Lab-Tek™) for microscopic inspection. Next day, the cells were transfected with Fugen HD as described above and incubated at 37°C in a 5 % CO2 atmosphere. The pictures were taken 24 h later directly, without washing, to demonstrate intracellular localization and distribution of the fluorescent proteins and fusion proteins (FPs) as well as the apoptotic and dead cells using a Leica TCS SP5 microscope. The optical slice thickness was 700 nm. The excitation wave-length of 543 nm was used to detect fluorescence signals (553-670 nm with a maximum at 610 nm). To increase the contrast of the optical sections, 12-20 single exposures were averaged. The image acquisition parameters were adapted to show signal intensities in accordance with the visible microscopic image.
Results & Discussion
It is well known that ROS is able to damage and finally kill cells. Our first intention was to clarify whether different FPs producing different amounts of ROS kill the host cells after illumination with normal daylight. Using the proteins (Green and Yellow or Red which achieved the maximal cell killing) we intended to investigate the influence of the intracellular localization on this cell killing effect. Therefore we first compared the survival of EGFP, EYFP, KRED expressing HeLa cells after illumination with white light. In this first attempt the cell survival was related to the tested FPs. The graphs indicate a different decrease of the cell number (cell tightness) expressed as a percentage depending on the time course of the illumination (Figure 7), also shown in Table 2 as counted colonies. The amount of HeLa cells stably transfected with pEGFP showed a slight decrease from 158 to 148 after 120 min illumination; during the illumination time up to 120 min the cell number was consistent with the control's cell number. The HeLa cells transfected with pEYFP featured a higher sensitivity against daylight illumination. Already a decrease from 162 cells after 30 minutes illumination time to 121 cells after 180 minutes illumination was observed. HeLa cells transfected with pKRED exhibited a clear linear decrease of the cell number from 155 to 103 from 30 up to 180 minutes illumination time course.
The graph demonstrates the influence of the illumination time on the cellular phenotype and displays the relative number of morphologically intact HeLa cells.
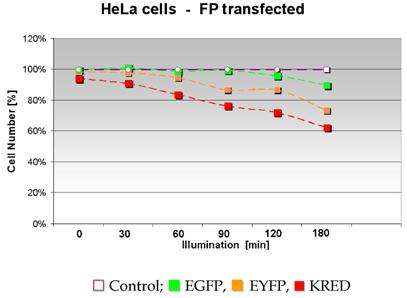
All cells with the different FPs were illuminated for the given time periods and counted. The control is set to 100%.
| 0 | 30 | 60 | 90 | 120 | 180 | [min] | |
|---|---|---|---|---|---|---|---|
| 165 | 165 | 165 | 165 | 165 | 165 | Control | |
| 165 | 167 | 163 | 164 | 158 | 148 | HeLa - EGFP | |
| 162 | 161 | 156 | 143 | 143 | 121 | HeLa - EYFP | |
| 155 | 150 | 138 | 126 | 119 | 103 | HeLa - KRED |
In the next experiments we focused on the influence of the intracellular localization of KRED.
The quantitative difference maybe influenced by differences in the absorptions coefficients, spectral inhomogeneity of the incident light or in different ROS building capacities and should be subject of further investigations. Here we investigated the cytotoxicity of the photodynamic effects caused by KillerRed and especially by its fusion proteins like the KRED-mem (Figure 8) locating the FPs to membrane, the KRED-Lamin B1 (Figure 9) variant with location to the nuclear surrounding lamin structure, and the histone H2A-KRED located in the chromatin structure (Figure 10). The CLSM pictures show clearly the detected KRED.
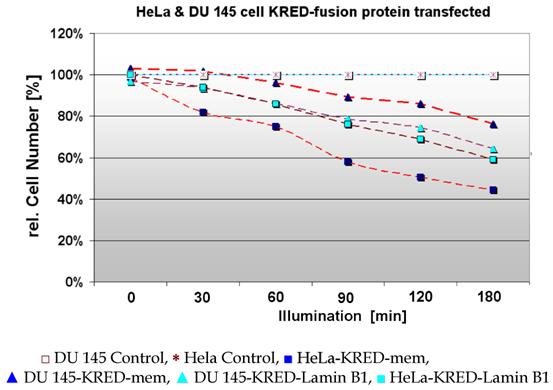
For a time course in cell killing we used HeLa and DU 145 cells both stably transfected with the above mentioned protein expressing KRED constructs. Cell survival was calculated by the decrease of the cell number expressing different FPs after illumination for increasing time periods measured. The current cell numbers are listed in Figure 11 and in Table 2.
The percentage value corresponding to the cell numbers of the different cell lines is exhibited.
| 0 | 30 | 60 | 90 | 120 | 180 | [min] | |
|---|---|---|---|---|---|---|---|
| 160 | 160 | 160 | 160 | 160 | 160 | DU 145 Control | |
| 160 | 160 | 160 | 160 | 160 | 160 | HeLa Control | |
| 165 | 163 | 154 | 143 | 138 | 122 | DU 145-KRED-mem | |
| 158 | 131 | 120 | 93 | 81 | 71 | HeLa-KRED-mem | |
| 155 | 150 | 138 | 126 | 119 | 103 | DU 145-KRED-Lamin | |
| 160 | 149 | 137 | 121 | 125 | 98 | HeLa-KRED-Lamin | |
| 158 | Incapable to measure | DU 145-H2A-KRED | |||||
| 160 | Incapable to measure | HeLa-H2A-KRED | |||||
In Table 2 the impact of the local position of KRED on the viability of two different eukaryotic tumor cell lines HeLa and DU 145 is described. Values for cell lines with histone H2A-KRED are missing.
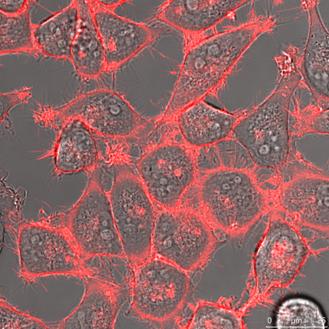
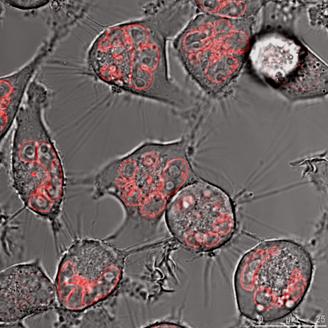
HeLa cells (left) and DU 145 (right) stably transfected with pKillerRed-mem vector encoding membrane-targeted KillerRed. The bars represent 25 µm (bottom right corner).
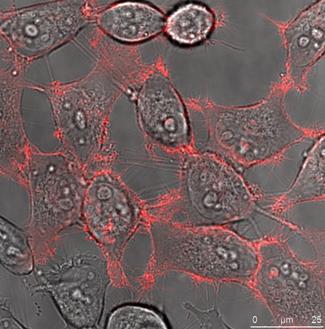
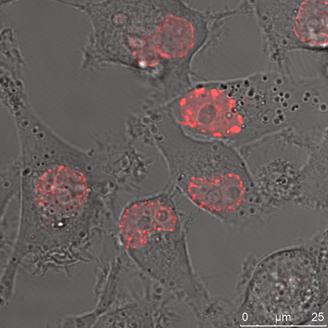
HeLa cells (left) and DU 145 cells (right) stably transfected with the pKillerRed Lamin B1 vector expressing the fusion protein KRED-Lamin B1. The bars represent 25 µm (bottom right corner)
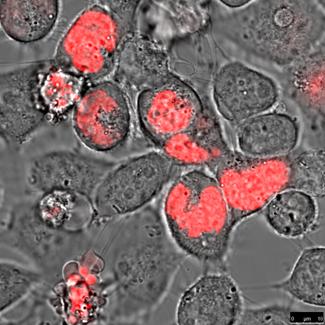
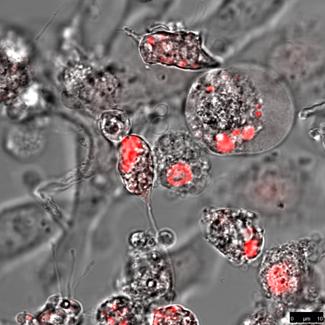
HeLa cells (left) and DU 145 cells (right) stably transfected with the pH2A-KillerRed vector expressing the fusion protein histone H2A-KRED located in the cell nuclei. The bars represent 10 µm (bottom right corner).
HeLa and DU 145 cells stably transfected with pKRED-mem and pKRED-Lamin to different cellular locations. Morphologically intact cells were counted after the illumination's time points: 30, 60, 90, 120, 180 minutes.
Despite several attempts we could not establish cell lines with histone H2A-KRED fusion proteins. Therefore we had to use transiently transfected cells instead and we investigated these.
An estimation of the cell number of both investigated cell lines; transfected with the histone H2A-KRED fusion protein was also not possible. However the intracellular localization of the histone H2A-KRED fusion protein could be scrutinized as visualized in Figure 10. Here we could detect apoptosis-typical morphologies in the cell nuclei of both cell lines, (structural changes such as half-moon-, sikle-, horseshoe-shaped nuclei, and chromatin condensation with the observation of seemingly “empty” red fluorescence areas in the nuclei. Both, the HeLa and the DU 145 cells show clearly formations of apoptotic membrane blebs, a typical sign of apoptosis and necrosis as a response to caspase activation as documented by the Barros Okada group22.
With increasing duration of light exposure of HeLa cells expressing the KRED-Lamin B1 protein, the cell number was continuously reduced from 160, 149, 137, 121, 125, and 98 measured from 30 to 180 minutes respectively. Under identical illumination conditions the decrease of the cell number of DU 145 cells expressing the fused KRED-Lamin B1 protein is slightly lower but in phase with the HeLa cell number's decrease (155, 150, 138, 126, 119, and 103 measured at identical time points as above) (Table 2). Both tested cell lines feature a similar sensitivity after illumination against the tested KRED located with Lamin B1. The picture of the pKRED-mem transfected cell lines is completely different: Indeed, Table 2 displays continuous decreases of the cell number of DU 145 cells from 165 to 122 indicating a lower sensitivity against the membrane-attached KRED protein in comparison to the cell's sensitivity for KRED-Lamin B1. Additionally, an extreme daylight sensitivity of KRED-mem expressing HeLa cells is evident as revealed by the strongly decreased cell numbers (158 to 71), listed in Table 2. The different behaviour of the tested cell lines with KRED located in subcellular membranes can be explained by different cell toxicity.
The observed higher sensitivity of the HeLa cells against KRED-mem as well as the KRED-Lamin B1 protein could be possibly explained by a higher ROS-cytotoxicity originated from the different spatial distribution patterns. A different physical proximity to intracellular membranes of the fusion proteins could cause ROS-overloading overcoming cell immanent detoxification systems. Furthermore recent data suggest a conjunction with DNA index and sensitivity against different cell toxic agents. The DU 145 cell line is deemed to an appropriate candidate intractable against different therapeutic interventions, which can be constituted by its aneuploid karyotype responsible for the contumaciousness.
The fact that the subcellular KRED's localization is pivotal for cytotoxic effects becomes apparent after transfection of HeLa and DU 145 cells with the plasmid encoding the H2A-KRED histone H2A fusion protein. After white light illumination the transfected cell lines except the control cells stopped the proliferation. Measurements of morphologically intact cells were therefore not possible (see Table 2).
Additionally, a dramatic cell killing effect emphasizes the critical role of the undamaged chromatin for intact living cells. Oxidative stress induced by increased intracellular ROS formation leads to different necrotic and apoptotic processes23. The involved cellular caspase induction and the NFκβ mechanisms are discussed24. But despite all documented intrinsic cell mechanisms protecting against ROS-damages the targeting and the enrichment result in a high local concentration of the KRED in the chromatin of tumor cells circumventing all cell-immanent protective barriers and sufficient for cell killing after exposure to daylight.
Outlook
The FP variant, called “KillerRed” (KRED) isolated by the Lukyanov group, acts as a photosensitizer. The high potential of the ROS produced by KRED in cell killing after exposition to daylight should be considered as an interesting tool for clinical use in the photodynamic therapy (PDT) of tumors. However we are aware that a successful molecule in PDT must fulfil manifold interrelated requirements, like intracellular accumulation after incorporation, and the ability of retention in the target neoplastic cell specifity. Furthermore questions of general features of photodynamic therapy of tumors in this context must be answered in relation to the normally used efficient photosensitizing dyes, like acridines, porphyrins, psoralens and xanthenes. It would be promising, if a delivery and it's retention of significant amounts of KRED could succeed into the target tissue. However the physical laws cannot be circumvented and the inappropriate gradient into the skin allows a depth of daylight penetration of few millimeters only and therefore the PDT focuses on epidermal tumors like skin cancer. Also an important aspect is the impact of the pivotal role of ROS, intracellularly generated, on the control of the oxygen-regulated genes, associated with cellular hypoxic stress response, like the HIF gene family25,26. Further effects e.g. ras-induction, and as aforementioned, NFκβ which should be discussed and considered as encouraged by Perkel in The-Scientist (http://www.the-scientist.com/blog/display/23010/) under the topic “KillerRed: The Hypoxia Connexion”
Additionally FP-imaging-based cell death studies depending on the intracellular localization of the particular ROS-expressing reporter proteins can contribute to a better understanding of the highly complex gene expression network.
Acknowledgements
We like to thank Harald Hermann for the provided Lamin B1 sequence.
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Szent-Gyorgyi A. The living state and cancer. Physiol Chem. Phys. 1980;12(2):99-110
2. Velando A, Torres R, Alonso-Alvarez C. Avoiding bad genes: oxidatively damaged DNA in germ line and mate choice. Bioessays. 2008;30(11-12):1212-1219
3. Mayr M, Sidibe A, Zampetaki A. The paradox of hypoxic and oxidative stress in atherosclerosis. J. Am. Coll. Cardiol. 2008;51(13):1266-1267
4. Puddu P, Puddu GM, Cravero E, Rosati M, Muscari A. The molecular sources of reactive oxygen species in hypertension. Blood Press. 2008;17(2):70-77
5. Voeikov VL. Reactive oxygen species--(ROS) pathogens or sources of vital energy? Part 1. ROS in normal and pathologic physiology of living systems. J Altern Complement Med. 2006;12(2):111-118
6. Davies KJ. Oxidative stress: the paradox of aerobic life. Biochem Soc Symp. 1995;61:1-31
7. Touyz RM. Reactive oxygen species as mediators of calcium signaling by angiotensin II: implications in vascular physiology and pathophysiology. Antioxid. Redox. Signal. 2005;7(9-10):1302-1314
8. Davies KJ. An overview of oxidative stress. IUBMB. Life. 2000;50(4-5):241-244
9. Filomeni G, Rotilio G, Ciriolo MR. Cell signalling and the glutathione redox system. Biochem. Pharmacol. 2002;64(5-6):1057-1064
10. Greabu M, Battino M, Mohora M, Olinescu R, Totan A, Didilescu A. Oxygen, a paradoxical element? Rom. J. Intern. Med. 2008;46(2):125-135
11. Jimenez-Banzo A, Nonell S, Hofkens J, Flors C. Singlet oxygen photosensitization by EGFP and its chromophore HBDI. Biophys. J. 2008;94(1):168-172
12. Greenbaum L, Rothmann C, Lavie R, Malik Z. Green fluorescent protein photobleaching: a model for protein damage by endogenous and exogenous singlet oxygen. Biol. Chem. 2000;381(12):1251-1258
13. Muller-Taubenberger A, Anderson KI. Recent advances using green and red fluorescent protein variants. Appl. Microbiol. Biotechnol. 2007;77(1):1-12
14. Bou-Abdallah F, Chasteen ND, Lesser MP. Quenching of superoxide radicals by green fluorescent protein. Biochim. Biophys. Acta. 2006;1760(11):1690-1695
15. Dixit R, Cyr R. Cell damage and reactive oxygen species production induced by fluorescence microscopy: effect on mitosis and guidelines for non-invasive fluorescence microscopy. Plant J. 2003;36(2):280-290
16. Singh K, Kang PJ, Park HO. The Rho5 GTPase is necessary for oxidant-induced cell death in budding yeast. Proc. Natl. Acad. Sci. U. S. A. 2008;105(5):1522-1527
17. Hadj Amor IY, Smaoui K, Chaabene I, Mabrouk I, Djemal L, Elleuch H, Allouche M, Mokdad-Gargouri R, Gargouri A. Human p53 induces cell death and downregulates thioredoxin expression in Saccharomyces cerevisiae. FEMS Yeast Res. 2008;8(8):1254-1262
18. Bulina ME, Chudakov DM, Britanova OV, Yanushevich YG, Staroverov DB, Chepurnykh TV, Merzlyak EM, Shkrob MA, Lukyanov S, Lukyanov KA. A genetically encoded photosensitizer. Nat. Biotechnol. 2006;24(1):95-99
19. Shagin DA, Barsova EV, Yanushevich YG, Fradkov AF, Lukyanov KA, Labas YA, Semenova TN, Ugalde JA, Meyers A, Nunez JM, Widder EA, Lukyanov SA, Matz MV. GFP-like proteins as ubiquitous metazoan superfamily: evolution of functional features and structural complexity. Mol. Biol. Evol. 2004;21(5):841-850
20. Bulina ME, Lukyanov KA, Britanova OV, Onichtchouk D, Lukyanov S, Chudakov DM. Chromophore-assisted light inactivation (CALI) using the phototoxic fluorescent protein KillerRed. Nat. Protoc. 2006;1(2):947-953
21. Gerthsen C, Kneser H, Vogel H. Strahlungsenergie. Berlin: Springer. 1977
22. Barros LF, Kanaseki T, Sabirov R, Morishima S, Castro J, Bittner CX, Maeno E, Ando-Akatsuka Y, Okada Y. Apoptotic and necrotic blebs in epithelial cells display similar neck diameters but different kinase dependency. Cell Death. Differ. 2003;10(6):687-697
23. Lemasters JJ, Qian T, Elmore SP, Trost LC, Nishimura Y, Herman B, Bradham CA, Brenner DA, Nieminen AL. Confocal microscopy of the mitochondrial permeability transition in necrotic cell killing, apoptosis and autophagy. Biofactors. 1998;8(3-4):283-285
24. Bubici C, Papa S, Pham CG, Zazzeroni F, Franzoso G. NF-kappaB and JNK: an intricate affair. Cell Cycle. 2004;3(12):1524-1529
25. Kaelin WGJr. ROS: really involved in oxygen sensing. Cell Metab. 2005;1(6):357-358
26. Chandel NS, Maltepe E, Goldwasser E, Mathieu CE, Simon MC, Schumacker PT. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc. Natl. Acad. Sci. U. S. A. 1998;95(20):11715-11720
Author contact
![]() Correspondence to: Dr. Klaus Braun, German Cancer Research Center (DKFZ), Dept. of Medical Physics in Radiology, Im Neuenheimer Feld 280, D-69120 Heidelberg, Germany. Tel: +49 6221 42 2495; Fax: +49 6221 42 3326. k.braunde
Correspondence to: Dr. Klaus Braun, German Cancer Research Center (DKFZ), Dept. of Medical Physics in Radiology, Im Neuenheimer Feld 280, D-69120 Heidelberg, Germany. Tel: +49 6221 42 2495; Fax: +49 6221 42 3326. k.braunde

 Global reach, higher impact
Global reach, higher impact