Impact Factor
ISSN: 1449-1907
Int J Med Sci 2005; 2(4):122-128. doi:10.7150/ijms.2.122 This issue Cite
Research Paper
Differential gene expression in HIV/SIV-associated and spontaneous lymphomas
1. Institute of Molecular Genetics, Russian Academy of Sciences, Kurchatov sq. 2, Moscow, Russia
2. National Research Center of Hematology, Russian Academy of Medical Sciences, Novozykovskii pr. 4a, Moscow, Russia
3. Department of Infection Pathology, German Primate Center, Kelnerweg, 4, Goettingen, Germany
4. Department of Virology and Immunology, German Primate Center, Kelnerweg, 4, Goettingen, Germany
Received 2005-6-2; Accepted 2005-8-29; Published 2005-10-1
Abstract
Diffuse large B-cell lymphoma (DLBCL) is more prevalent and more often fatal in HIV-infected patients and SIV-infected monkeys compared to immune-competent individuals. Molecular, biological, and immunological data indicate that virus-associated lymphomagenesis is similar in both infected hosts. To find genes specifically overexpressed in HIV/SIV-associated and non-HIV/SIV-associated DLBCL we compared gene expression profiles of HIV/SIV-related and non-HIV-related lymphomas using subtractive hybridization and Northern blot analysis. Our experimental approach allowed us to detect two genes (a-myb and pub) upregulated solely in HIV/SIV-associated DLBCLs potentially involved in virus-specific lymphomagenesis in human and monkey. Downregulation of the pub gene was observed in all non-HIV-associated lymphomas investigated. In addition, we have found genes upregulated in both non-HIV- and HIV-associated lymphomas. Among those were genes both with known (set, ND4, SMG-1) and unknown functions. In summary, we have demonstrated that simultaneous transcriptional upregulation of at least two genes (a-myb and pub) was specific for AIDS-associated lymphomas.
Keywords: non-Hodgkin's lymphoma, diffuse large B-cell lymphoma, HIV/SIV-associated lymphomas, spontaneous, differentially expressed genes, subtractive hybridization
1. INTRODUCTION
Lymphoid neoplasms represent a heterogeneous group of malignancies including Hodgkin's disease, non-Hodgkin lymphomas (NHLs), various leukemias, and multiple myeloma. In recent years the molecular mechanism of lymphomagenesis has been studied intensively. The formation and progression of B-cell NHLs affect 3.5-12% of patients including diffuse large B-cell lymphomas (DLBCL) infected with the human immunodeficiency virus (HIV) [1, 2]. In individuals with AIDS DLBCL is 60 to 200-fold more likely to occur than in general population [3, 4]. Infection with simian immunodeficiency virus (SIV) in some monkeys also leads to B-cell NHLs pathologically and clinically similar to those of HIV-infected patients [5-8]. SIV‑associated NHLs are therefore an appropriate model to study the role of immunodeficiency virus in lymphomagenesis.
Molecular studies have revealed both similarities and differences between HIV-associated and non-HIV-associated lymphomas [9-11]. Although overexpression of some genes in a large proportion of HIV-associated DLBCL as compared to spontaneous DLBCL has been reported [9], specific differences in gene expression have not yet been detected [11]. Thus the question whether unique mechanisms leading to HIV-associated NHLs do exist remains open.
Recently, using PCR-based two-step subtractive hybridization we identified spectra of genes overexpressed in human HIV-associated lymphomas [12] and monkey SIV-associated lymphomas [13] as compared with B-lymphocytes from blood and lymph nodes of healthy individuals. To reveal the difference in gene expression and to find genes both up- and down-regulated during the formation of the lymphomas, we performed subtractive hybridization with centroblastic and immunoblastic HIV-associated DLBCLs in both directions. Transcription levels of the genes overexpressed in HIV/SIV-associated lymphomas were compared with those in human spontaneous lymphomas. The data obtained have revealed a specific difference in the expression pattern of several genes in HIV/SIV-associated as compared to non-HIV-associated (spontaneous) DLBCLs.
2. METHODS
Tumor tissue and cells
Biopsy specimens from two lymphomas (h1 and h2) from HIV-1-infected AIDS-patients (males, age 43 and 36, respectively) were kindly provided by Prof. Dr. I. Schedel, Medical School, Hannover, Germany [12]. The material from lymphoma h1 was taken from the left tonsil. The specimens from lymphoma h2 were taken from the liver hilus. The latter patient was classified as WR-6 stage of AIDS. The tumors were B-cell NHL of centroblastic type (lymphoma h1) and immunoblastic type (lymphoma h2). Cells from both tumors harbored Epstein-Barr virus genomes and contained EBER-1 and EBNA-2 mRNAs [14]. Three rhesus monkeys (M. mulatta) #1725, #7198 and #1153 (m1, m2, and m3, respectively) developed B-NHL after infection with SIVmac251 [13]. All samples were examined histologically and tissue blocks were trimmed to exclude areas of surrounding nonlymphoid tissues. Lymphoma biopsy specimens were stored at –80ºC. Using RT-PCR, we analyzed the expression levels of the three oncogenes (bcl-2, bcl-6 and c-myc) and two suppressor gene (Rb и p53) both in the human and monkey AIDS-related lymphomas. The expression profile of these genes was similar to that found in normal human B-lymphocytes (data not shown).
Spontaneous non-HIV-associated lymphoma biopsy specimens were provided by the Hematology Scientific Center of the Russian Academy of Medical Sciences. The characteristics of these tumors are summarized in Table 1.
Human and monkey B-lymphocytes were isolated with LymphoSep (ICN Biomedicals) from peripheral blood of healthy donors and monkeys.
Characteristics of human spontaneous lymphomas
| ## | Subtype of lymphoma (REAL classification) | Sex/Date of birth |
|---|---|---|
| #3 (# 3860-864/01) | DLBCL | woman/1900 |
| #4 (# 956-998/01) | FL, stage II, mixed cells | woman/1945 |
| #5 (# 3681-3682/01) | FL | man/1955 |
| #7 (# 1638-1644/00) | DLBCL | woman/? |
| #8 (# 2300-1766/01) | DLBCL | man/1961 |
| #9 (# 1212-1216/01) | FL, stage I, preferentially small cells | man/1936 |
| #10 (# 1537-1539) | DLBCL | man/1975 |
| #13 (# 334-336/01) | Nodular sclerotic HD | man/1971 |
| #14 (# 2218-2219/00) | Nodular sclerotic HD | man/1978 |
DLBCL – diffuse large B-cell lymphoma; FL – follicular lymphoma; HD – Hodgkin's disease
RNA extraction, labeling, and hybridization
Total cellular RNA was isolated from tissues and B lymphocytes dispersed in liquid nitrogen in the presence of 4 M guanidine isothiocyanate as described earlier [15]. RNA was extracted twice with phenol, and its concentration was determined spectrophotometrically. The quality of the isolated RNA was confirmed by a horizontal agarose gel electrophoresis as well as by OD 260/280 ratios.
The primers structure and the annealing temperatures used for PCR
| Gene | Primers | Annealing, t oС |
|---|---|---|
| β-actin | 5'-TGCTTCTAGGCGGACTATGAC-3' 5'-AGAAGTGGGGTGGCTTTTAGG-3' | 64 |
| set | 5'-ACCTGGTTTACTGACCATTCTGA-3' 5'-TCTTCTTCACCTTCATCCTCATC-3' | 59 |
| ND4 | 5'-TCCCCACCTTGGCTATCATC-3' 5'-TTGTTGGCTCAGGAGTTTGA-3' | 60 |
| a-myb | 5'-AAGAAGAATCAGGCACTCAACTG-3' 5'-GGAAGTGTTAGGGTTTGGTTTCT-3' | 60 |
| capn4 | 5'-CCACAAGCTTTTGTTCTCTCAGTA-3' 5'-CACAGGTACAGGGGAGAGGTTAC-3' | 60 |
| pub | 5'-TACTGACCGAGTGCTGAGACTACT-3' 5'-GCAATTTGCCATATCAATAAAGAA-3' | 60 |
Northern blot analysis was performed as described earlier [12]. Membranes with RNA were UV irradiated and hybridized with [32P]-labeled SalI-fragments of cDNA clones generated by subtraction combined with differential screening or with [32P]-labeled PCR-fragments of the corresponding genes. As a control, we used a [32P]-labeled β-actin PCR-amplification product. The nucleotide sequences of the primers are presented in Table 2. Dot-hybridization of the subtracted human cDNA library with radioactively labeled monkey cDNAs was performed as previously described [16]. cDNA and PCR-fragments were labeled by the random-prime method (Prime-a-Gene Labeling System, Promega, USA). [32P]dCTP was obtained from Amersham International (Amersham, UK). The radioactive bands were quantified by Phosphorimager analysis (Molecular Dynamics, USA).
Subtractive cloning
A PCR-based technique was performed. A detailed protocol how to generate cDNA libraries, isolation of lymphoma-specific cDNA by subtractive hybridization and the differential screening was published previously [13]. In the previous study [12] we used RNAs from lymphoma h1 and h2 cells as tracer and driver, respectively. In this study, RNA from lymphoma h1 cells was used as driver, and RNA from lymphoma h2 cells as tracer.
Sequencing of DNA and analysis
Cloned cDNAs were sequenced using an Amersham Quick-Denature-Plasmid-Sequencing Kit. A search for similarity of the subtracted sequences with known sequences was performed with the BLAST DataBase.
3. RESULTS
Different factors and a variety of genes may contribute to the chain of events that eventually lead to lymphomagenesis. In an initial study to identify genes differentially transcribed in HIV and SIV-associated DLBCLs we employed a two-step subtractive hybridization using the RNA from lymphoma cells of HIV/SIV-infected individuals as tracer and as driver the RNA from B lymphocytes of an uninfected human and monkey [12, 13]. However, it remained to be elucidated whether enhanced transcription of some genes in these lymphomas was associated with the malignant transformation of these cells or with another factors, such as different proliferation rates of the cell populations examined. Another open question is whether there are differences in the expression of genes in HIV/SIV-related and spontaneous lymphomas.
To address these questions, the subtraction hybridization between the two human HIV-related lymphomas (h1 and h2) cDNAs was performed in both directions: 1) the cDNA population of lymphoma h1 as tracer, and the cDNA population of lymphoma h2 as driver; and vice versa, 2) the cDNA population of lymphoma h2 as tracer, and the cDNA population of lymphoma h1 as driver.
The cDNAs selected by the two-step differential screening were sequenced and compared with nucleotide sequences available in BLAST Databases. Partial preliminary data concerning the first subtraction were published [12]. The complete results of two subtractive hybridizations in both directions are given in Table 3.
A comparison of their sequences allowed us to subdivide the cDNAs into two groups. The first group includes cDNAs selected both by subtractive hybridization between the two lymphomas (Table 3) and between lymphomas and B-lymphocytes [12] (the set oncogene, constant part of the λIg gene, the mitochondrial genes of NADH dehydrogenase subunit 4 (ND4), the interferon-inducible gene 6-16 (Inf-ind), the 16S rRNA gene (most probably the humanin gene [17]). These results confirmed the adequacy of our method and suggested that the use of RNA from B-lymphocytes was quite applicable for the detection of B‑cell lymphomas specific gene expression.
cDNA nucleotide sequences selected by two-step hybridization and subsequent two-step differential screening of cDNA populations from lymphoma h1 and h2 as overexpressed both in each lymphoma in comparison with the other lymphoma and human normal B-lymphocytes
| HIV-associated DLBCL | The differentially transcribed genes | NCBI Acc N |
|---|---|---|
| h1 | set | M93651 |
| a-myb | X66087 | |
| IL4R | X54425 | |
| IL5 | BG599356 | |
| capn4 | X04106 | |
| ND4 | V00662 | |
| TAP2 | U07844 | |
| HnRNP A1 | BG644331 | |
| 16S rRNA (humanin) | AY011166 | |
| IFN-ind | U22970/X14583 | |
| λIg | X14583 | |
| γIg | M63438 | |
| KIAA1536 | AB040969 | |
| DKFZp547I094 | AK024405 | |
| FLJ20554 | AK000561 | |
| FLJ23277 | AK026930 | |
| EST | BG599355 | |
| EST | BG599357 | |
| EST | BG599358 | |
| EST | BG599359 | |
| h2 | SMG-1 | CB252001 |
| ribosomal protein S8 | CB252002 | |
| KIAA1350 | AB037771 | |
| FLJ20306 | NM017756 | |
| FLJ39323 fis | АК096642 | |
| FLJ14117 | BC038668 | |
| FLJ30053 fis | АК054615 | |
| EST | CB252000 |
capn4 – the calpain subunit 4 gene, ND4 – the NADH dehydrogenase subunit 4 gene, IL5 – the interleukin 5 gene, λIg – the λ-chain of immunoglobulin gene, γIg – the γ-chain of immunoglobulin gene, IFN-ind – the interferone-inducible gene 6-16, IL4R – the interleukin 4 receptor gene, TAP2 – the transport protein gene, hnRNP A1 – the human ribonucleoprotein А1 gene; SMG-1 – the phosphatidylinositol kinase (PIK)-related kinase 1 gene, EST – expressed sequenced tags
The second group represents those cDNAs that were only revealed by subtractive hybridization between two HIV-related lymphomas. In this group of upregulated genes there were the a-myb oncogene, the interleukin 4 receptor gene (IL4R), the gene of the transporter protein TAP2, the gene of protease calpain 4 small subunit (capn4) and other (in case of lymphoma h1) and SMG-1, the gene of ribosomal protein S8 (in case of lymphoma h2), as well as several genes of unknown function (9 and 8 in the case of lymphoma h1 and h2, respectively). The latter genes may represent new genes associated with lymphomagenesis but undetectable by microarray.
Differences in expression of genes of the second group might be due to different origin and molecular mechanisms acting in these two types of human HIV-associated DLBCL. Perhaps the subtraction performed would hardly shed light on the role of HIV in the development of lymphomas, and it would be better to subtract HIV-associated DLBCL cDNAs from those of spontaneous DLBCL. But in earlier experiments, such a difference was not detected [11]. We have suggested that at least some of the genes preferentially expressed in one of these lymphomas might be involved in HIV-associated lymphomagenesis, and this suggestion was confirmed.
We found earlier that some genes (set, COX-II) highly expressed in one of DLBCL, as compared to B-cells, were actually upregulated in both HIV-associated lymphomas [12]. The expression of several genes isolated with subtractive hybridization between h1 and h2 (Fig 1, see also [12]) was evaluated using Northern blot analysis in both human lymphomas and human B-lymphocytes. The expression of the a-myb oncogene was shown to be higher (about 5 times) in lymphoma h1 (lane h1) than in lymphoma h2 (lane h2), but in both cases higher (about 5-10 times) than in human B-lymphocytes (lane B) (Fig. 1) when normalized by β-actin hybridization to these filters). Likewise, the expression levels of the SMG-1 and capn4 genes in both lymphomas were also higher (about 2-3 times) than those in normal B‑lymphocytes.
Northern blot analysis of differential transcription in human HIV-associated lymphomas h1 and h2, monkey SIV-associated lymphomas m1, m2, m3, and human normal B-lymphocytes. 32P‑labeled PCR-fragments of the a-myb oncogene or the SMG-1 gene were hybridized to RNA from human normal B-lymphocytes (lane B), human HIV-associated lymphomas h1 (lane h1) and h2 (lane h2), monkey SIV-associated lymphomas m1 (lane m1), m2 (lane m2), and m3 (lane m3). Rehybridization with a 32P-labeled PCR-fragment of β-actin gene was used as control (bottom)
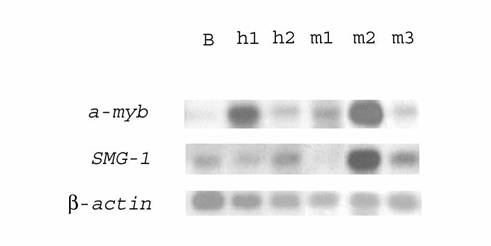
Macaques infected with SIV are an appropriated animal model for HIV infection and AIDS of humans [5-8, 13]. We supposed that some genes were overexpressed both in HIV- and SIV-associated lymphomas. Using dot and blot hybridization, transcription of genes upregulated in human HIV-associated lymphoma was studied in three SIV‑associated monkey lymphomas and monkey B-lymphocytes. To this end, about 100 cDNAs from subtracted human cDNA libraries of lymphomas h1 and h2 were analyzed by dot blot hybridization with [32P]‑labeled cDNA populations from SIV-associated monkey lymphomas and monkey B‑lymphocytes. Those cDNAs whose hybridization signals were markedly stronger with lymphoma cDNA than with cDNA of B-lymphocytes were further analyzed by Northern blot hybridization. Some genes (a-myb, set, SMG-1, ND4) involved in HIV-associated lymphomagenesis were overexpressed in one or more SIV-associated lymphomas (Tables 4, 5). Their transcription in other SIV-associated lymphomas was unchanged or even downregulated. For example, Fig. 1 shows that the transcription of the SMG-1 gene was 8 fold upregulated in lymphoma m2, unchanged in lymphoma m3 and even downregulated (no expression) in lymphoma m1 (lanes m1, m2, and m3). The a-myb oncogene was about 2.5-7 fold overexpressed in all SIV-associated monkey lymphomas (Fig. 1, lanes m1, m2, and m3). However, the capn4 gene was not transcribed in SIV-associated lymphomas. The results obtained were in accord with our earlier results of Northern blot hybridization with SIV-associated monkey mRNA [13].
Earlier we identified the pub gene as upregulated in SIV-associated monkey DLBCL [13]. The pub gene (also known as KIAA0129 or TRIM14) was previously found to be expressed in the human myeloid cell line KG-1 [18]. Northern blotting with RNAs from SIV-associated monkey lymphomas [13] demonstrated overexpression of the pub gene in the cells of all three SIV-associated monkey lymphomas as compared to B-lymphocytes. Northern blot hybridization of a PCR-fragment of pub with RNA from human HIV-associated lymphomas h1 and h2 and B-lymphocytes revealed increased levels of this gene transcription in both these human lymphomas (in lymphoma h1 higher than in h2) [16]. The two genes (a‑myb and pub) were thus overexpressed both in human HIV-associated and monkey SIV‑associated lymphomas and seemed to be common for virus-specific lymphomagenesis in human and monkey.
Selected human HIV-associated lymphomas h1 and h2 genes overexpressed in SIV-associated monkey lymphomas in comparison with monkey B lymphocytes (results of the blot- and dot-hybridization)
| Genes | NCBI Acc N |
|---|---|
| set | M93651 |
| a-myb | X66087 |
| IL4R | X54425 |
| ND4 | V00662 |
| pub | D50919 |
| TAP2 | U07844 |
| HnRNP A1 | BG644331 |
| λIg | X14583 |
| KIAA1536 | AB040969 |
| DKFZp547I094 | AK024405 |
| FLJ23277 | AK026930 |
| EST | CB252000 |
| EST | CB252001 |
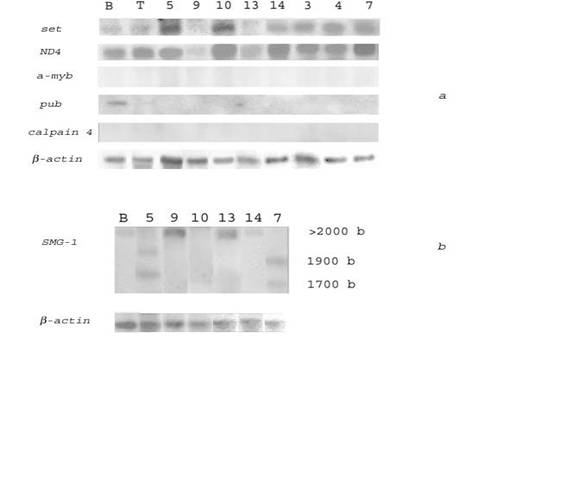
To find genes specifically overexpressed in HIV/SIV-associated and/or non-HIV/SIV-associated DLBCLs, we compared transcription levels of the selected genes in HIV/SIV-associated and human non-HIV-associated (spontaneous) lymphomas. Northern blot hybridization of several genes with RNAs from 9 spontaneous human lymphomas (4 - DLBCL, 3 - follicular lymphomas (FL), 2 - Hodgkin's disease (HD)) is presented in Fig. 2a, 2b and Table 5.
Northern blot analysis of the transcription levels of the set, ND4, and SMG-1 genes in human spontaneous lymphomas and normal B- and T-lymphocytes. (a) 32P-labeled PCR‑fragments of the set and a-myb oncogenes, and ND4, pub and capn4 genes were hybridized to RNA from human normal B-lymphocytes (lane B), T-lymphocytes (lane T), human spontaneous DLBCLs # 3, 7, 10 (lanes 3, 7, 10), FL # 4, 5, 9 (lanes 4, 5, 9), HD # 13, 14 (lanes 13, 14). (b) A 32P-labeled SalG1-fragment of a cDNA clone homologous to the SMG-1 gene was hybridized to RNA from human normal B-lymphocytes (lane B), human spontaneous DLBCLs # 7, 10 (lanes 7, 10), FL # 5, 9 (lanes 5, 9), HD # 13, 14 (lanes 13, 14). Rehybridization with a 32P-labeled PCR‑fragment of β-actin gene was used as control (bottom)
Summary of gene expression levels in human non-HIV-associated and HIV/SIV-associated lymphomas (in comparison with normal B-lymphocytes)
| Lymphomas | The genes upregulated only in virus-associated lymphomas | The genes upregulated in virus-associated and spontaneous lymphomas | |||||
|---|---|---|---|---|---|---|---|
| a-myb | pub | capn4 | set | ND4 | SMG1 | ||
| HIV-associated | h1 (DLBCL) | ++ | + | ++ | ++ | ++ | + |
| h2 (DLBCL) | + | + | + | + | + | ++ | |
| SIV-associated | m1 (DLBCL) | + | ++ | N | + | + | - |
| m2 (DLBCL) | +++ | ++ | N | N | + | ++ | |
| m3 (DLBCL) | + | + | N | + | + | + | |
| non-HIV-associated | #3 (DLBCL) | N | - | N | + | + | N |
| #7 (DLBCL) | N | - | N | + | + | + | |
| #8 (DLBCL) | N | - | N | + | + | N | |
| #10 (DLBCL) | N | - | N | + | + | - | |
| #4 (FL) | N | - | N | + | + | + | |
| #5 (FL) | N | - | N | + | N | + | |
| #9 (FL) | N | - | N | N | - | + | |
| #13 (HD) | N | - | N | N | N | + | |
| #14 (HD) | N | - | N | + | + | N | |
“+”, “++” – upregulation, “-” – downregulation, “N” – no changes, DLBCL – diffuse large B-cell lymphoma, FL – follicular lymphoma, HD – Hodgkin's desease, EST (NCBI Acc N CB252001)
The results indicated that the set oncogene was transcribed 2-6 times more abundantly in several non-HIV-associated lymphomas (as compared with normal human B‑lymphocytes) including all DLBCLs (# 3, 7, 8, 10), some FLs (# 4, 5) and HDs (# 14). The gene of the mitochondrial NADH dehydrogenase subunit 4 (ND4) was 2-4 times overexpressed in all analyzed spontaneous DLBCLs (# 3, 7, 8, 10), and in some FL (# 4) and HD (# 14). The SMG-1 gene was highly transcribed in DLBCL # 7, in FLs # 5, 9 and in HDs # 13, 14. In lymphomas # 5 and 7 SMG-1 transcripts of different length were formed (possible this is result of alternative splicing of mRNA). However, Northern blot analysis revealed no transcription of the a-myb and capn4 genes in non-HIV-associated lymphomas and B-lymphocytes (Fig. 2a). The pub gene was slightly transcribed in B‑lymphocytes and not transcribed in all non-HIV-associated lymphomas (Fig. 2a).
Thus, three genes (a-myb, pub, capn4) are upregulated exclusively in HIV-associated DLBCLs and probably specific for these lymphomas. Two of them (a-myb, pub) were found upregulated also in SIV-associated DLBCLs. The pub gene was downregulated in all non-HIV-associated lymphomas analyzed. Also, we revealed a set of genes (set, ND4, SMG-1) which upregulated both in non-HIV-associated and in HIV/SIV-associated lymphomas.
4. DISCUSSION
Our studies were aimed at the detection of new genes involved in the process of lymphomagenesis. Numerous approaches have been proposed to identify and analyze genes differentially expressed in cancer cells and particularly in lymphomas [9, 11-13, 19-22]. These techniques allowed to detect sets of genes up- or downregulated in malignant cells used as diagnostic markers to characterize different types of lymphomas. The cDNA microarray technology [19-22] has allowed the investigation of global gene expression profiles in cancer. Although cDNA microarray is a powerful tool for the identification of differentially expressed genes, this methodology has several potential limitations [22].
To identify genes differentially expressed in HIV/SIV-associated lymphomas, we used the PCR-based two-step subtractive hybridization. This method does not need any previously cloned cDNA sets and allows to detect unknown genes. However, it remained to be elucidated whether enhanced transcription of some genes in lymphomas detected by this approach was associated with malignant transformation or with other factors, e.g. different proliferation rates of the cell populations examined. To answer this question, we performed subtractive hybridization between DLBCLs from two different AIDS-patients. The results of several independent experiments demonstrated that many of the genes revealed by us previously were also upregulated in the two lymphoma types. In HIV-associated lymphomas h1 and h2, some upregulated cDNA clones were found to be homologous to known genes including the set and a-myb oncogenes, genes of ND4, IL4R, IL5, SMG-1, ribosomal protein S8, immunoglobulins, ribonucleoprotein hnRNP A1, transport protein ТАР2, CAPN4, etc. In addition, 17 overexpressed clones homologous to expressed sequence tags (ESTs) were detected. They may represent novel genes whose transcription is upregulated in lymphomas. A number of the genes detected in these experiments have not been previously associated with DLBCL of different origin.
A comparison of expression profiles of some of these genes with those for the genes revealed earlier by subtractive hybridization in SIV-associated lymphomas [13] were very similar (Tables 4 and 5). About 10 genes were found to be overexpressed both in HIV- and SIV-related lymphomas.
To find genes specific for HIV/SIV-associated and/or non-HIV/SIV-associated DLBCLs, we examined the expression levels of these 10 upregulated genes in spontaneous lymphomas. The results obtained (Table 5) allowed us to subdivide these genes into two groups. Firstly, we detected three genes (a-myb, pub, capn4) upregulated exclusively in human HIV-associated DLBCLs (and not in spontaneous lymphomas) and apparently specific for these DLBCLs. Transcription levels of the lymphoma-specific genes in three SIV-associated DLBCLs showed that two genes (a-myb, pub) of this group were overexpressed also in SIV-associated DLBCLs and might be involved in virus-specific lymphomagenesis in human and monkey. Secondly, the transcription of set, ND4, SMG-1 was increased in most HIV- and non-HIV-associated lymphomas. These genes may be involved in both HIV-associated and non-HIV-associated lymphomagenesis. Some genes of this group (set, SMG-1, ND4) were overexpressed at least in one SIV-associated lymphoma, in the other SIV-associated lymphomas investigated their expression was unchanged or even downregulated.
Moreover, we compared the expression of the selected lymphoma specific genes in normal human T‑lymphocytes and the Jurkat T-cell line. Expression of set, SMG-1, and ND4 was unchanged, and pub and a-myb were not transcribed in normal human T‑lymphocytes and the Jurkat cells (data not shown). These results suggested an association of the former genes exclusively with B cell but not T cell lymphomas.
Attempts to reveal genes overexpressed in HIV-related DLBCL versus DLBCL have already been reported [9, 11]. Preliminary evidence for the high and specific expression of the TCL-1 proto-oncogene in HIV-related lymphomas [9] was confirmed only partially [11]. In contrast to our data, genes specifically expressed in AIDS-related lymphomas were not detected. This contradiction is most likely explained by technical differences. For example, Patrone et al. [11] arbitrarily excluded some apparently “uninteresting” genes. But we have shown earlier that genes like ATP synthase, cytochrome b, cytochrome c oxidase, and 16S rRNA are specifically upregulated in lymphomas [12, 13]. Also, the gene named 16S RNA most probably relates to the humanin gene [17], since its transcript contains poly(A).
According to our data (Tab. 5) a-myb was overexpressed in all AIDS-related lymphomas. At the same time, transcription of a-myb is unchanged or even reduced in all non-AIDS-related lymphomas analyzed here. a-myb overexpressed in many cancer cells and in several germinal center (GC) B-like DLBCL [19, 23]. Among the other genes (bcl-6, bcl-7A) the a-myb defined subtype GC B-like DLBCL but it was not a general rule. The a-myb product is known to upregulate bcl-2 in various lymphomas and to maintain the expression of c-myc in mouse B cell lymphoma [24]. Both these gene upregulated mainly in subtype activated B-like DLBCL [19]. Using RT-PCR, we analyzed the expression levels of the three oncogenes (bcl-2, bcl-6 and c-myc) both in the human and monkey AIDS-related lymphomas. The expression profile of these genes was similar to that found in normal human B-lymphocytes. This suggested that AIDS-related lymphomas have been hardly referred to any definite subtype of DLBCL.
Some of the genes we identified have not been implicated previously in DLBCL. According to our data the pub gene, also known as KIAA0129 or TRIM14, was overexpressed only in AIDS-related DLBCL, whereas its transcription was downregulated in all non-HIV-associated lymphomas (Table 5). No reports on transcription levels of pub gene in lymphomas were previously available. This gene was earlier identified in the human myeloid KG-1 cell line [18]. Recently, it was shown, that pub is predominantly transcribed in hematopoietic tissues and inhibits the activity of the PU.1 transcription factor important for B-lymphocytes differentiation and proliferation [25]. Our data suggest that the pub gene simultaneously with a-myb may be involved in malignant transformation of B cells in HIV/SIV-associated DLBCL.
We have shown that the gene coding for the small subunit of calcium-dependent cysteine proteinase calpain 4 (capn4) is upregulated in both HIV-associated lymphomas, but not SIV-associated lymphomas. The role of calpains in apoptosis was studied almost exclusively. However, calpain inhibits apoptosis in acute lymphoblastic leukemia cells and might be associated with malignization of these cells [26]. Recent studies of unusual effects of calpains in cancer cells [27-29] suggest that calpain participates in cell malignization by digesting the p53 tumor suppressor. Besides, calpains participate in induction of HIV replication [30]. However, there are no data on overexpression of capn4 in spontaneous DLBCLs. These and our data suggest that capn4 is a candidate gene specific for human HIV-associated lymphomas.
We have also detected a high level transcription of set (TAF-1, template activating factor-1) in both HIV-associated and non-HIV-associated lymphoma tissues. The SET protein is highly homologous to NAP (nucleosome assembly protein). Alternative splicing of set mRNA leads to the formation of at least two protein forms, TAF-1α and TAF-1β [31], the latter being the SET protein. SET/TAF-1β is a member of the INHAT (inhibitor of histone acetyltransferase) complex. SET is also an inhibitor of protein phosphatase 2A [32]. Inhibition of this phosphatase induces an increase in telomerase activity known to be involved in cell immortalization [33]. We detected an enhanced transcription of the set gene (the only 780 bp long mRNA coding for SET/TAF-1β) both in HIV/SIV-associated and several spontaneous human lymphomas. Microchip studies also revealed that set was highly expressed in human spontaneous DLBCLs [19]. Thus enhanced expression set might be associated with the malignisation of germinal center-derived large B cells of different origin.
The human HIV-associated and monkey SIV-associated lymphomas analyzed overexpressed a limited set of mitochondrial genes of the oxidative phosphorylation pathway [12,13]. We demonstrated changes in the expression of ND4 mitochondrial genes in various lymphomas. It is difficult to discriminate whether this phenomenon is a cause or a consequence of oncogenesis. But our experiments with immortalized fibroblasts, used as a model of early stage of oncogenesis, suggest that enhanced transcription of mitochondrial genes can be associated with the early stages of oncogenesis [34].
The SMG-1 kinase gene was upregulated in one human HIV-associated, one monkey SIV‑associated and several human non-HIV-related lymphomas. Previously, this gene was not associated with lymphomas development. The SMG-1 kinase is a member of the phosphatidylinositol kinase (PIK)-related kinase family involved in nonsense-mediated mRNA decay [35]. The defective SMG-1 kinase may contribute to the process of malignant transformation in B-cell lymphomas of different origin.
In conclusion, using subtractive hybridization we detected three genes (a-myb, pub and capn4) upregulated predominantly in HIV-associated DLBCLs. So far, the only known gene presumably specific for HIV-associated immunoblastiс plasmacytoid lymphomas was TCL1 [9]. The study the role of the immunodeficiency virus in lymphomagenesis, we measured the transcription levels of human lymphoma-specific genes in SIV-associated DLBCLs thought to be an appropriate model. Two of the three genes (a-myb, pub) upregulated in HIV-related lymphomas were overexpressed in SIV-associated lymphomas and might be simultaneous involved in virus-specific lymphomagenesis in man and monkey. All genes detected might be specifically upregulated in HIV-associated DLBCLs and these are targets for therapy of these lymphomas. The set, ND4 and SMG‑1genes involved in both spontaneous and HIV/SIV-associated lymphomagenesis. The latter two genes have not been implicated previously in DLBCL.
Abbreviations
HIV: human immunodeficiency virus; SIV: simian immunodeficiency virus; DLBCL: diffuse large B-cell lymphoma; NHL: non-Hodgkin's lymphoma; AIDS: acquired immunodeficiency syndrome; FL: follicular lymphoma; HD: Hodgkin's disease; capn4: the calpain subunit 4 gene; ND4: the NADH dehydrogenase subunit 4 gene; IL5: the interleukin 5 gene; λIg: the λ-chain of immunoglobulin gene; γIg: the γ-chain of immunoglobulin gene; IFN-ind: the interferon-inducible gene 6-16; IL4R: the interleukin 4 receptor gene; TAP2: the transport protein gene; hnRNP A1: the human ribonucleoprotein А1 gene; SMG-1 : the phosphatidylinositol kinase (PIK)-related kinase 1 gene; EST : expressed sequenced tags
Acknowledgements
This work was supported by the Russian Ministry of Education and Science and the Russian Foundation for Basic Research (grant 04-04-49420).
Conflict of interests
The authors have declared that no conflict of interest exists.
References
1. Cote TR, Biggar RJ, Rosenberg PS. et al. Non-Hodgkin's lymphoma among people with AIDS: incidence, presentation and public health burden. AIDS/Cancer Study Group. Int J Cancer. 1997 ;73:645-650
2. Frisch M, Biggar RJ, Engels EA, Goedert JJ. Association of Cancer with AIDS-related immunosuppression in adults. J Am Med Assoc (JAMA). 2001 ;285:1736-1745
3. Weiss RA. Viruses, cancer and AIDS. FEMS Immunol Med Microbiol. 1999 ;26:227-232
4. Knowles DM, Pirog EC. Pathology of AIDS-related lymphomas and other AIDS-defining neoplasms. Eur J Cancer. 2001 ;37:1236-1250
5. Feichtinger H, Li SL, Kaaya E. et al. A monkey model for Epstein Barr virus-associated lymphomagenesis in human acquired immunodeficiency syndrome. J Exp Med. 1992 ;176:281-286
6. Rezikyan S, Kaaya EE, Ekman M. et al. B-cell lymphomagenesis in SIV-immunosuppressed cynomolgus monkeys. Int J Cancer. 1995 ;61:574-579
7. Hannig H, Matz-Rensing K, Kuhn EM. et al. Cytokine gene transcription in SIV and HIV-associated non-Hodgkin's lymphomas. AIDS Res Hum Retrovir. 1997 ;13:1589-1596
8. Maggiorella MT, Monardo F, Koanga-Mogtomo ML. et al. Detection of infectious simian immunodeficiency virus in B- and T-cell lymphomas of experimentally infected macaques. Blood. 1998 ;91:3103-3111
9. Teitell M, Damore MA, Sulur GG. et al. TCL1 oncogene expression in AIDS-related lymphomas and lymphoid tissues. Proc Nat Acad Sci U.S. A. 1999 ;96:9809-9814
10. Breen EC, Boscardin WJ, Detels R. et al. Non-Hodgkin's B cell lymphoma in persons with acquired immunodeficiency syndrome is associated with increased serum levels of IL10, or the IL10 promoter -592 C/C genotype. Clin Immunol. 2003 ;109:119-129
11. Patrone L, Henson SE, Teodorovic J. et al. Gene expression patterns in AIDS versus non-AIDS-related diffuse large B-cell lymphoma. Exp Mol Pathol. 2003 ;74:129-139
12. Tarantul VZ, Nikolaev AI, Hannig H. et al. Detection of Abundantly Transcribed Genes and Gene Translocation in Human Immunodeficiency Virus Associated non-Hodgkin's Lymphoma. Neoplasia. 2001 ;2:132-142
13. Tarantul VZ, Nikolaev AI, Martynenko A. et al. Differential gene expression in B-cell non-Hodgkin's lymphoma of SIV-infected monkey. AIDS Res Hum Retrovir. 2000 ;16:173-179
14. Pingel S, Hannig H, Motz-Rensing K. et al. Detection of Epstein-Barr virus small RNAs EBER1 and EBER2 in lymphomas of SIV-infected rhesus monkeys by in situ hybridisation. Int J Cancer. 1997 ;72:160-164
15. Sambrook J, Fritch EF, Maniatis T. Molecular cloning: A laboratory manual. USA: Cold Spring Harbor Laboratory Press. 1989
16. Nenasheva VV, Maksimov VV, Nikolaev AI, Tarantul VZ. Comparative analysis of the level of gene transcription in two types of HIV-associated lymphoma. Mol Gen Mikrobiol Virusol (Moscow). 2001 ;4:27-31
17. Maximov V, Martynenko A, Hunsmann G, Tarantul V. Mitochondrial 16S rRNA gene encodes a functional peptide, a potential drug for Alzheimer's disease and target for cancer therapy. Medical Hypotheses. 2002 ;59:670-673
18. Nagase T, Seki N, Tanaka A. et al. Prediction of the coding sequences of unidentified human genes. IV. The coding sequences of 40 new genes (KIAA0121-KIAA0160) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res. 1995 ;2:167-174
19. Alizadeh AA, Eisen MB, Davis RE. et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000 ;403:503-511
20. Alizadeh AA, Staudt LM. Genomic-scale gene expression profiling of normal and malignant immune cells. Current Opinion in Immunology. 2000 ;12:219-225
21. Wellmann A, Thieblemont C, Pittaluga S. et al. Detection of differentially expressed genes in lymphomas using cDNA arrays: identification of clusterin as a new diagnostic marker for anaplastic large-cell lymphomas. Blood. 2000 ;96:398-404
22. Monti S, Savage KJ, Kutok JL. et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood. 2005 ;105:1851-1861
23. Heckman CA, Mehew JW, Ying GG. et al. A-Myb up-regulates Bcl-2 through a Cdx binding site in t(14;18) lymphoma cells. J Biol Chem. 2000 ;275:6499-6508
24. Arsura M, Hofmann CS, Golay J. et al. A-myb rescues murine B-cell lymphomas from IgM-receptor-mediated apoptosis through c-myc transcriptional regulation. Blood. 2000 ;96:1013-1020
25. Hirose S, Nishizumi H, Sakano H. Pub, a novel PU.1 binding protein, regulates the transcriptional activity of PU.1. Biochem Biophys Res Commun. 2003 ;311:351-360
26. Zhu DM, Uckun FM. Calpain inhibitor II induces caspase-dependent apoptosis in human acute lymphoblastic leukemia and non-Hodgkin's lymphoma cells as well as some solid tumor cells. Clin Cancer Res. 2000 ;6:2456-2463
27. Kurland JF, Meyn RE. Protease inhibitors restore radiation-induced apoptosis to Bcl-2-expressing lymphoma cells. Int J Cancer. 2001 ;96:327-333
28. Wang XD, Rosales JL, Magliocco A. et al. Cyclin E in breast tumors is cleaved into its low molecular weight forms by calpain. Oncogene. 2003 ;22:769-774
29. Witkowski JM, Zmuda-Trzebiatowska E, Swiercz JM. et al. Modulation of the activity of calcium-activated neutral proteases (calpains) in chronic lymphocytic leukemia (B-CLL) cells. Blood. 2002 ;100:1802-1809
30. Teranishi F, Liu ZQ, Kunimatsu M. et al. Calpain is involved in the HIV replication from the latently infected OM10.1 cells. Biochem Biophys Res Commun. 2003 ;303:940-946
31. Nagata K, Kawase H, Handa H. et al. Replication factor encoded by a putative oncogene, set, associated with myeloid leukemogenesis. Proc Nat Acad Sci. 1995 ;92:4279-4283
32. Li H, Zhao LL, Funder JW, Liu JP. Protein phosphatase 2A inhibits nuclear telomerase activity in human breast cancer cells. J Biol Chem. 1997 ;272:16729-16732
33. Li M, Makkinje A, Damuni Z. The myeloid leukemia-associated protein SET is a potent inhibitor of protein phosphatase 2A. J Biol Chem. 1996 ;271:11059-11062
34. Nenasheva VV, Nikolaev AI, Dubovaya VI. et al. Analysis of expression of a series of lymphoma-specific genes in human fibroblasts immortalized by SV40 virus. Mol Biol (Moscow). 2004 ;38:265-275
35. Denning G, Jamieson L, Maquat LE. et al. Cloning of a novel phosphatidylinositol kinase-related kinase. Characterization of the human SMG-1 RNA surveillance protein. J Biol Chem. 2001 ;276:22709-22714
Author contact
![]() Corresponding address:
Corresponding address:
Prof. V.Z. Tarantul, Deputy Director, Institute of Molecular Genetics RAS, Kurchatov Sq., 2, Moscow 123182, Russia. Phone/Fax: +7-095-196-00-02; Fax: +7-095-196-02-21; tarantulras.ru

 Global reach, higher impact
Global reach, higher impact