ISSN: 1449-1907International Journal of Medical Sciences
Int J Med Sci 2014; 11(8):788-795. doi:10.7150/ijms.8417 This issue Cite
Research Paper
Spatiotemporal Expression of HDAC2 during the Postnatal Development of the Rat Hippocampus
1. Children Nutrition Research Center, Children's Hospital of Chongqing Medical University, Chongqing 400014, China;
2. Ministry of Education Key Laboratory of Child Development and Disorders, Chongqing Key Laboratory of Translational Medical Research in Cognitive Development and Learning and Memory Disorders, Chongqing 400014, China.
Abstract
Background: Histone acetylation, which is a chromatin modification of histone tails, can dynamically regulate the expression of various genes in normal development. HDAC2 is a negative regulatory factor of acetylation and closely related to learning and memory. NSE is a nerve marker and vital for maintaining physiological functions in nervous system. Currently, few studies associated with the expression pattern of HDAC2 in postnatal rat hippocampus have been reported. This study aimed to explore the temporal and spatial expression pattern of HDAC2, helping to reveal the expression characteristics of HDAC2 during postnatal neuronal maturation. Materials and Methods: With NSE as a biomarker of neuronal maturation at postnatal days 1, 3, 7 and weeks 2, 4, and 8 (P1D, P3D, P7D, P2W, P4W, P8W), the expression patterns of HDAC2 in rat hippocampus were examined using real-time PCR and western blotting. Additionally, the subcellular distribution of HDAC2 was analysed by immunofluorescence. Results: We found that HDAC2 was highly expressed in the neonatal period and decreased gradually. HDAC2 expression was widely distributed in neurons of hippocampal CA1, CA3 and DG regions and gradually shifted from the nucleus to the cytoplasm during postnatal development. Altogether, the expression of HDAC2 decreased gradually with different subcellular localizations throughout development. Conclusions: The observed results indicate that the expression levels of HDAC2 become lower and with different subcellular localizations in neurons during hippocampal neuronal maturation, suggesting the specific expression characteristics of HDAC2 might play an important role during postnatal learning-memory function and development.
Keywords: HDAC2, NSE, hippocampus, development, epigenetic.
Introduction
The hippocampus plays a crucial role in the generation of episodic and declarative memory during the maturation of neurons as well as during spatial learning and in the formation of memory. The hippocampus is responsible for the proper formation and function of the postnatal brain development in rodents [1]. It has been reported that epigenetic mechanisms are involved in the control of the accessibility of chromatin to transcriptional factors in neurons. These transcription factors can in turn impact gene expression and lead to modulation of neurotransmission, synaptic plasticity, and disease states [2]. Histone acetylation involves post-translational modification, such as the removal or addition of an acetyl group to specific lysine residues in the amino-terminus of histone residues by histone deacetylases (HDACs) and histone acetytransferases (HATs). Non-selective HDAC inhibitors (HDACi) can improve deficiencies in synaptic plasticity and cognition-related behaviours in the animal models of neurodegenerative diseases and clinical treatments [2-9]. However, currently little is known about the actual HDAC isoforms and the mechanisms mediating histone acetylation during disease progression. Thus, the identification of specific HDAC isoforms is critical for the application of HDACi in the therapeutic treatment of cognitive impairment.
HDACs are classified into four types. HDAC2 is a member of the class I HDACs. Some findings have demonstrated that HDAC2 is an important negative regulator of memory formation [10]. However, to date the characteristic of HDAC2 during postnatal development is not well explored. Few studies have addressed a complete pattern and timeline of dynamic changes in expression levels, localisation or the multiple functional areas of HDAC2 in the period from birth to sexual maturity [11-13]. Because changes in HDAC2 expression can reflect the physiological needs of different stages of development, systematic study of the expression pattern of HDAC2 is very necessary. Neuron-specific enolase (NSE) is considered to be a marker of neurons and neuroendocrine cells and can reflect the characteristics of the nervous system [14-16]. Therefore, our current study tried to utilise NSE as a biomarker of neuronal maturation in the hippocampus to analyse the HDAC2 expression pattern underlying changes during the postnatal development period.
Materials and methods
Animals
Wistar rats were obtained from the Experimental Animal Centre of Chongqing Medical University, China [certificate: SCXK (Yu) 2007~0001]. All rats were housed under specified pathogen free (SPF) laboratory conditions. The rats were housed in individual stainless steel cages at a constant temperature of 19°C-25°C with 55%-65% relative humidity; artificial daylight was put on between 7 a.m. and 7 p.m., and food and water were available ad libitum. The day of birth was designated as P0. All animal experimental protocols were approved by the Chongqing Medical University Institutional Animal Care and Use committee. All efforts were made to minimize the number of animals used and alleviate their suffering.
Real-time PCR of HDAC2 and NSE of the hippocampus
Extraction of RNA of hippocampal tissue was performed using the total RNA isolation system (Takara, Japan). The mRNA was reverse transcribed into cDNA using the PrimeScript RT Reagent Kit (Takara, Japan). Real-time PCR reactions were performed using a RealMasterMix Kit (Takara, Japan) and a Real-time PCR instrument (Bio-Rad, USA). The reaction protocol was carried out as follows: 95 °C ×30 s, 95 °C ×5 s and 60 °C ×20 s for 40 cycles, and followed by 60 °C for 30 s. Melt curve analysis was utilised when the temperature was brought gradually up to 95 °C. The data were represented as gene expression and were calculated using the ratio of the relative concentration of the target gene to that of GAPDH with the same samples by CFX Manager Software. Primer sequences for HDAC2, NSE and GAPDH were designed with the Primer 5 software as follows.
HDAC2, forward: 5'-GCTGCTTCAACCTAACTG-3'; reverse: 5'-CTCATACGTCCAACATCG-3'
NSE, forward: 5′-CTGTTTGCTGCTCAAGGTC-3′; reverse: 5′-TCCCACTACGAGGTCTGC-3′
GAPDH, forward: 5′-CCTGGAGAAACCTGCCAAG-3′; reverse: 5′-CACAGGAGACAACCTGGTCC-3′
Western blotting of HDAC2 and NSE of the hippocampus
Hippocampal tissue was prepared for protein extraction with RIPA buffer (Bioteke, China). Protein concentrations in the homogenates were determined with the micro BCA protein assay kit (Bioteke, China) with an enzyme-labelling instrument (Thermo varioskan flash, USA). Approximately 40 μg of total protein samples were loaded per lane onto a 10% SDS-polyacrylamide gel (Beyotime, China). Proteins were transferred to 0.45 μm polyvinylidene fluoride membranes (Millipore, USA) after electrophoretic separation. All membranes were then blocked with 5% skim milk in TBST at 37 °C for 1 h and probed with anti- NSE (at dilution of 1/2000) (Abcam, UK), anti-HDAC2 (at dilution of 1/1500) (Millipore, USA), and anti-β-actin (at dilution of 1/800) (Santa Cruz, USA) primary antibodies at 4°C overnight. The proteins were probed with HRP-conjugated secondary antibodies (at dilution of 1/5000) (Santa Cruz, USA) at room temperature for 1 h and visualised using an enhanced chemiluminescence kit (Millipore, USA) with an ECL Imaging System (SynGene GBOX, UK). Band intensities were quantified using ImageJ software.
Immunofluorescence staining
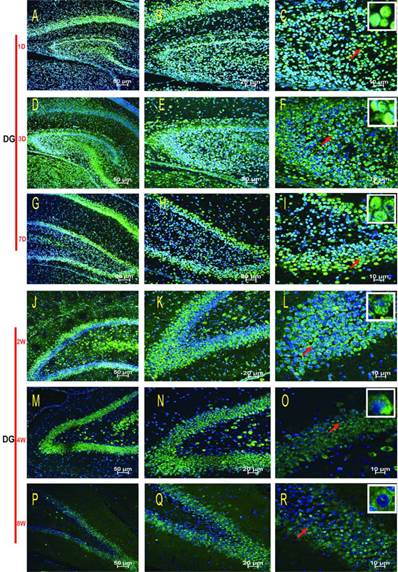
Animals were deeply anesthetised and perfused transcardially with 200 ml of ice-cold PBS (0.01 M) followed by 200 ml paraformaldehyde (4% in 0.1 M PBS). Brains were rapidly removed from the skull and post-fixed overnight in the same fixative at 4°C. Every sixth section of every brain was mounted onto slides for staining. After dehydration and paraffin embedding, the brains were sliced into coronal hemisections, with a 4.5 µm in thickness using the vibratome (Leica, Germany) and the sections were kept at 4°C in slice boxes before use. After permeabilisation (PBS 1X / Triton 2%, 10 min) and antigen retrieval (boiling citric acid / sodium citrate buffer, pH 6.0, 45 min), the hippocampus slices were blocked (5% goat serum, 30 min), followed by incubation with anti-HDAC2 antibody (at dilution of 1/100) (Millipore, USA), at 4°C overnight. After three washes (PBS 1X/Triton 0.1%), the hippocampus slices were incubated with dylight488-conjugated secondary antibodies (at dilution of 1/1000) (Jackson ImmunoResearch, USA) at room temperature for 1 hour. Additionally, nuclei were stained using 4'6'-diamidino-2-phenylindole dihydrochloride (DAPI) (at dilution of 1/2000) (Sigma, USA) for 10 min. Fluorescent images and fluorescence intensity were acquired with an Immunofluorescence Laser Confocal (Nikon, Japan). Control IgG of corresponding primary antibodies were used as negative controls. Three regions (CA1, CA3, DG) of the hippocanpus were all selected for each time point for statistical analyses of fluorescence expression intensity. Analysis was performed on the samples with the magnification of 400× with scale bars of 10 μm using NIS-Elements AR 3.1 software (NIKON, Japan). The different scales of HDAC2 expression shown in Table 1 were based on the subcellular localizations of HDAC2 in the three regions (CA1, CA3, DG) of the IF pictures with the magnification of right row 400× (C, F, I, L, O, R) with scale bars of 10 μm.
Subcelluar translocation.
| CA1 | CA3 | DG | ||||
|---|---|---|---|---|---|---|
| Cyto plasm | Nucleus | Cytoplasm | Nucleus | Cytoplasm | Nucleus | |
| 1D | -+ | +++ | -+ | +++ | -+ | +++ |
| 3D | -+ | +++ | -+ | ++ | + | +++ |
| 7D | -+ | +++ | + | ++ | + | +++ |
| 2W | + | ++ | ++ | + | + | ++ |
| 4W | ++ | + | ++ | + | + | ++ |
| 8W | +++ | + | +++ | + | ++ | + |
+++ : high expression, ++ : intermediate expression, + : low expression, -+ : micro expression.
Statistical analysis
The results were expressed as the means ± S.E.M. Statistical analyses were performed using one-way analysis of variance (ANOVA) or Student-Newman-Keuls test (SNK). The data were analysed using GraphPad Prism 5.0 software. p ≤ 0.05 was considered to be significant.
Results
Expression pattern of NSE in the hippocampus
First, we measured the mRNA expression levels of NSE in the rat hippocampus at postnatal 1, 3, and 7 D (P1D, P3D, P7D) and at 2, 4, and 8 W (P2W, P4W, P8W). From the data shown in Fig. 1A, we found that the mRNA expressions of NSE in the hippocampus tissue gradually increased with the growth and development of pups, and there were significant differences among 6 groups (#p<0.001, one-way ANOVA). The levels of NSE mRNA expression significantly increased from P3D to P4W, but there was no significant difference between P4W and P8W (ns, p≥0.05, SNK). Developmental regulation of NSE expression in the rat hippocampus was almost like an “s”, relatively steady in the beginning and end but with a sharp increase in the middle. Meanwhile, the changes in NSE protein levels were completely consistent with those of NSE mRNA expressions among 6 time points (Fig. 1B, 1C) (#p<0.001, one-way ANOVA). The expression levels of NSE protein increased significantly from the P3D to P4W, and the levels of NSE protein expression at P4W were near to those of P8W (ns, p≥0.05, SNK).
The regulation of NSE mRNA and protein expression levels in the rat hippocampus during postnatal development. The regulation of NSE mRNA (A) and protein (B) expression at the six time points (P1D, P3D, P7D, P2W, P4W, P8W) of postnatal development. Quantitative analysis of NSE protein expression levels (C). The values are the mean ± SEM. There are three samples for each time point and each sample was tested in triplicate independently. The mRNA was normalised to GAPDH. The protein samples were equally loaded and normalised to β-actin (#p<0.001 by one-way ANOVA and *p<0.05, **p<0.01, ***p<0.001 by SNK).
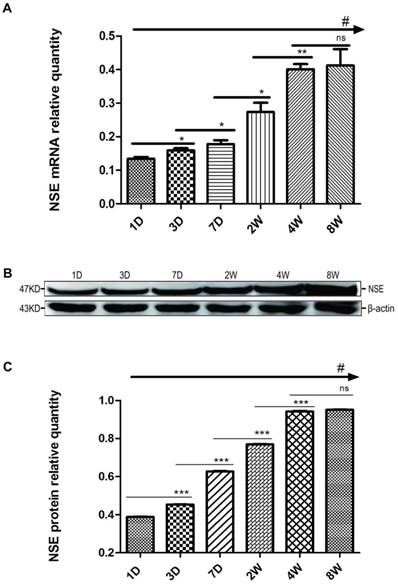
Changes in HDAC2 mRNA and protein expression in the rat hippocampus during postnatal development. The expression pattern of HDAC2 mRNA (A) and protein (B) expression at the six time points (P1D, P3D, P7D, P2W, P4W, P8W) of postnatal development. The mRNA was normalised to GAPDH. The protein samples were equally loaded and normalised to β-actin. Quantification analysis of protein expression levels of HDAC2 (C) in the hippocampus during the postnatal development. The values are displayed as the mean ± SEM. There are three samples for each time point and each sample was tested in triplicate independently (#p<0.001 by one-way ANOVA and *p<0.05, **p<0.01, ***p<0.001 by SNK).
Expression pattern of HDAC2 in the hippocampus
We also examined the expression profiles of HDAC2 in the hippocampus at postnatal 1, 3, and 7 D and at 2, 4, and 8 W. As shown in the Fig. 2A, the expression levels of HDAC2 mRNA in the hippocampus gradually decreased with the growth and development of pups, and there were significant differences among the 6 groups (#p<0.001, one way-ANOVA). The levels of HDAC2 mRNA expression slightly decreased from P1D to P3D with no significant difference between P1D and P3D (ns, p≥0.01, SNK). HDAC2 mRNA expression levels significantly decreased from P7D to P4W and remained similar between P4W and P8W (ns, p≥0.05, SNK). Contrasting with NSE, the mRNA regulatory pattern of HDAC2 was almost like a “trans-s”, relatively stable in the beginning and end but with a sharp decrease in the middle. Next, the changes of HDAC2 protein expression were surprisingly similar to the expressions of HDAC2 mRNA at all of the time points (Fig. 2B, 2C) (#p<0.001, one way-ANOVA). The expression levels of HDAC2 protein also significantly decreased from P3D to P4W, but there was no obvious difference between P1D and P3D or between P4W and P8W. Generally, based on the mRNA expression of HDAC2 and NSE, we readily observed that the regulation of HDAC2 expression was opposite to that of NSE expression during the postnatal growth and development of pups.
Functional area and subcellular location of HDAC2 in the hippocampus
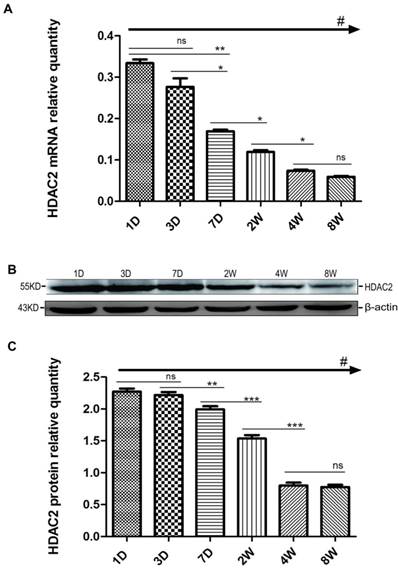
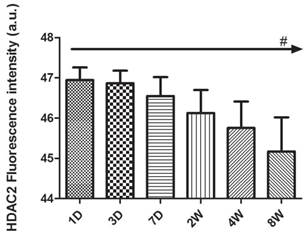
HDAC2 subcellular expression was analysed in the hippocampus by Immunofluorescence Laser Confocal Multi-magnification Scanning. HDAC2 expression was observed in the CA1 (Fig. 3), CA3 (Fig. 4) and DG (Fig. 5) regions of the hippocampus at P1, 3, and 7 D and at 2, 4, and 8W under 100×, 200×, and 400× scanning microscopy, respectively. HDAC2 was abundantly expressed in the neurons of the three regions of the hippocampus. Expression of HDAC2 also declined dynamically along with the development from P1D to P8W. The quantitative analysis of fluorescence intensity demonstrated that the levels of HDAC2 expression in hippocampus diminished from P1D to P8W (Fig. 6), which was consistent with the results of HDAC2 mRNA and protein expression exhibited above. Moreover, the changes in HDAC2 expression in the three regions of the hippocampus and translocation to cytoplasm from the nucleus throughout the postnatal development are shown in the white frames (Fig. 3, Fig. 4, Fig. 5, C, F, I, L, O, R) and in Table 1. We were surprised to find that HDAC2 expression localised both in the nucleus and cytoplasm of neurons rather than only in the nucleus. In the P1-7D period, little HDAC2 immunoreactivity was detected in the neuronal cytoplasm and was mostly located in the nucleus (Fig. 3, Fig. 4, Fig. 5, A-I, Table 1. P1D-7D). However, a significant amount of HDAC2 was expressed in the neuronal cytoplasm, and HDAC2 displayed weak expression in the nucleus in the P2-8W period (Fig. 3, Fig. 4, Fig. 5, J-R, Table 1. P2W-8W).
Discussion
The central nervous system in mammals, especially the hippocampus, undergoes significant postnatal development, which is particularly important for the maturation of neural function and is mainly responsible for learning and memory in adulthood. [17, 18]. NSE is a marker of mature neurons and can reflect the characteristics of the central nervous system during the development period well [14-16]. Therefore, to better understand the developmental changes in the hippocampus, we detected the temporally dynamic expression patterns of NSE in the postnatal hippocampus. We found that NSE increased gradually along the different stages and stayed steady in the adult stages. NSE expression is very abundant in adult brains [16], and our results verified this finding (Fig. 1). The dynamic changes in NSE imply the progressive maturation of hippocampal neurons and are reflected in our study based on the stable development of neurons in the rat hippocampus. Consequently, we can use NSE as a biomarker of neuronal maturation to observe the expression pattern of HDAC2 at the different stages of neuron development.
Subcellular localisation of HDAC2 in the CA1 region of the rat hippocampus during postnatal development. HDAC2 expression was readily detected in the neuronal nuclei at P1D-7D, and cytoplasmic expression was weak (A-I). The patterns of translocation to the neuronal cytoplasm from the nucleus were observed at P2W-8W (J-R). Red arrows indicate the location of the white frames, which display the respective cells from each time point. The magnification of the left row is 100× (A, D, G, J, M, P), middle row 200× (B, E, H, K, N, Q), and right row 400× (C, F, I, L, O, R); with scale bars of 50 μm, 20 μm, and 10 μm, respectively. Blue fluorescence: nucleus; Green fluorescence: HDAC2.
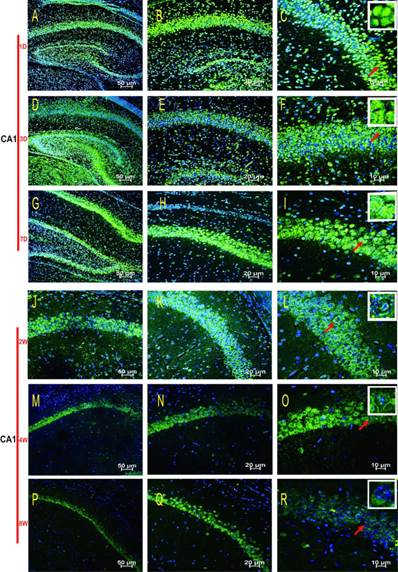
Subcellular localisation of HDAC2 in the CA3 region of the rat hippocampus during postnatal development. HDAC2 was mainly expressed in neuronal nuclei at the P1D-7D, and the cytoplasmic expressions were weak (A-I). The translocation expression pattern in the neuronal cytoplasm was observed at P2W-8W (J-R). Red arrows indicate the location of the white frames which show the respective cells at each time point. The magnification of the left row is 100× (A, D, G, J, M, P), middle row 200× (B, E, H, K, N, Q), and right row 400× (C, F, I, L, O, R) with scale bars of 50 μm, 20 μm, and 10 μm, respectively. Blue fluorescence: nucleus; Green fluorescence: HDAC2.
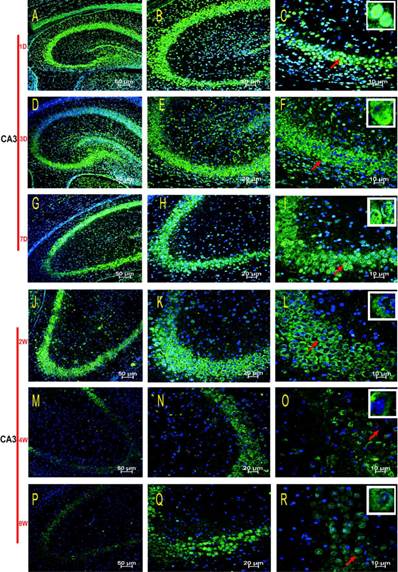
Subcellular localisation of HDAC2 in the DG region of the rat hippocampus during postnatal development. The HDAC2 immunoreactivity was located in the neuronal nuclei at P1D-2W, and the cytoplasmic expressions were weak (A-L). The expression pattern was translocation into the neuron cytoplasm during P4W-8W (M-R). Red arrows indicate the location of the white frames, which display the respective cells at each time point. The magnification of left row is 100× (A, D, G, J, M, P), middle row 200× (B, E, H, K, N, Q), and right row 400× (C, F, I, L, O, R) with scale bars of 50 μm, 20 μm, and 10 μm, respectively. Blue fluorescence: nucleus; Green fluorescence: HDAC2.
Quantification of fluorescence intensity in HDAC2 immunoreactivity of the rat hippocampus during postnatal development from P1D to P8W. Analysis was performed on the samples with the magnification of right row 400× (C, F, I, L, O, R) of Fig 3, 4, 5 with scale bars of 10 μm. The levels of HDAC2 protein expression peaked at P1D and then decreased gradually with postnatal development. The reported values are shown as the mean ± SEM, and the above data were confirmed in at least six samples for each time point and each sample was tested in triplicate (#p<0.001 by one-way ANOVA).
HDAC enzymes are present in animals, plants, and bacteria [19], and there are 11 isoforms of HDAC in the brain of the adult rat [20]. HDAC2 can negatively regulate the formation of hippocampus-dependent memory by decreasing synaptic plasticity and synapse number [21-26]. Some studies have demonstrated the distribution of HDAC2 in the central nervous system. Macdonald and Roskams showed the expression pattern of HDAC2 at embryonic day 13.5 (E13.5), postnatal day 7 (P7), and in the adult murine brain and reported predominant HDAC2 expression in neurons [11]. Yao's study demonstrated that HDAC2 was positively distributed in the hippocampal neurons of mice aged 3 months [12]. Yoo's research found that HDAC2 was abundantly expressed throughout the course of development in the cortex ranging from E12 to P9, maintained a similar expression level at all embryonic time points, and showed a two-fold decrease in postnatal mouse cerebellum [13]. However, the timing variation for HDAC2 throughout development from the early life period to sexual maturation in rat hippocampus was definitely unclear. The current study shows that HDAC2 exhibited a widespread distribution with relatively high levels in the CA1, CA3, and DG regions, especially at the neonatal stage in the rat hippocampus. Moreover, not only by morphological methods but also by systematic gene and protein studies, we revealed that the expression levels of HDAC2 decreased during early life development and kept at a stable and low level after P4W. When NSE was steadily expressed later, HDAC2 was also kept at a stable and low level after P4W. Some findings imply functional roles for HDAC2 in the maturation of neural cells [10, 11, 27-29], especially in migration and differentiation [13]. In addition, HDAC2 can affect basic excitatory neurotransmission [26, 29, 30]. Thus, the specific characteristics of HDAC2 expression may be one of the key elements of normal development of the hippocampus.
According to the expression patterns and the sequence homology relative to the yeast HDAC proteins, the mammalian HDAC enzymes are classified into four principal types. Class I (HDACs 1, 2, 3, and 8) proteins are thought to be located predominantly in the nucleus. Class IIa (HDACs 4, 5, 7, and 9) and Class IIb (HDACs 6 and 10) shuttle between the nucleus and the cytoplasm. Class III corresponds to the NAD+-dependent Sirtuin family. HDAC11 is a zinc-dependent deacetylase that has common features with class I and II HDACs and is the sole member of class IV so far [31-33]. However, we were surprised to find that HDAC2 also could shift from the nucleus to the cytoplasm similar to class II HDACs and other nuclear receptors [34, 35]. The HDAC2 expression patterns likely reflect the intrinsic characteristics in both the circuitry and function of these regions in hippocampus. The neurons that convey information within hippocampal circuits are the granule cells of the DG and the pyramidal cells of the CA3 and CA1 regions. CA1 is involved in spatial memory formation, and CA3 is involved more with pattern completion-based memory recall [36, 37]. DG is a novelty detector through its neuronal activation, giving salience to incoming sensory stimuli [38, 39]. The differences in HDAC2 expression over time may indicate that neurons are activated when diverse sensory stimuli or information flows at different developmental stages are challenged. In addition, neuronal histone acetylation is dynamically regulated in response to environmental stimuli that initiate memory formation [3]. The nucleo-cytoplasm shuttling of class IIa HDACs or receptors is due to transcriptional induction of genes for cellular adaptation with diverse stress signals by chromatin remodelling [38, 40]. HDAC2 expression declines and shifts from the nucleus to the cytoplasm gradually in our study, implying that functionally distinct roles are caused by abundant stimuli during postnatal development. In addition, the expression patterns of HDAC2 were similar across the different regions of the developing hippocampus. This result supports the hypothesis that HDAC2 plays important roles in mediating the distinct signals generated by region-specific genes, rather than the region-specific genes themselves. HDAC2 in distinct neural fates of different regions by diverse stimulus warrants further study. The enzymatic activity of HDACs can be regulated by protein kinases, phosphatases, other post-translational modifications or by protein-protein interaction [38, 40]. HDACs also deacetylate non-histone proteins. The gradual increase in histone acetylation can facilitate the ability of transcription factors to mediate gene expression for functional maturation. Moreover, the translocation of HDAC2 is speculated to be required for interaction with some necessary regulators. In this regard, HDAC2 is vital for maturation of neuronal function, possibly through some novel epigenetic regulator-mediated pathway. Certainly, our next task is to explore the exact roles of HDAC2 in neuronal or functional maturation and its specific relationship with NSE. Further studies of both the upstream and the downstream targets of HDAC2 will extend our knowledge of the biological role of HDAC2 which could lead to development of novel therapeutic strategies for cognitive damages. Taken together, spatiotemporal expression of HDAC2 shown in our data reinforces the notion that HDAC2 plays a critical, but extremely complicated role in neuronal maturation with specific developmental expression patterns. The findings connecting these changes to functional maturation, the underlying mechanisms of action, their direct substrates, and their biological roles warrant further examination.
In summary, the HDAC2 expression levels decreased gradually during early life development. Moreover, HDAC2 expression displayed different subcellular localizations in neurons during postnatal hippocampal development. These findings reveal that the specific expression characteristics of HDAC2 might play an important role during postnatal learning-memory function and development.
Abbreviations
HATs: histone acetytransferases; HDACs: histone deacetylases; HDAC2: histone deacetylase 2; NSE: neuron-specific enolase; DG: dentate gyrus.
Acknowledgements
This work was supported by the Specialized Research Fund for the Doctoral Program of Projects funded by Chinese Ministry of Education (No. 20115503110003).
Competing Interests
The authors have declared that no competing interest exists.
References
1. McCaffery P, Zhang J, Crandall JE. Retinoic acid signaling and function in the adult hippocampus. J Neurobiol. 2006;66:780-91
2. Kim MS, Akhtar MW, Adachi M. et al. An essential role for histone deacetylase 4 in synaptic plasticity and memory formation. J Neurosci. 2012;32:10879-86
3. Bahari-Javan S, Maddalena A, Kerimoglu C. et al. HDAC1 regulates fear extinction in mice. J Neurosci. 2012;32:5062-73
4. Chuang DM, Leng Y, Marinova Z, Kim HJ, Chiu CT. Multiple roles of HDAC inhibition in neurodegenerative conditions. Trends Neurosci. 2009;32:591-601
5. Kilgore M, Miller CA, Fass DM. et al. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer's disease. Neuropsychopharmacology. 2010;35:870-80
6. McQuown SC, Wood MA. HDAC3 and the molecular brake pad hypothesis. Neurobiol Learn Mem. 2011;96:27-34
7. Monsey MS, Ota KT, Akingbade IF, Hong ES, Schafe GE. Epigenetic alterations are critical for fear memory consolidation and synaptic plasticity in the lateral amygdala. PLoS One. 2011;6:e19958
8. Morris MJ, Karra AS, Monteggia LM. Histone deacetylases govern cellular mechanisms underlying behavioral and synaptic plasticity in the developing and adult brain. Behav Pharmacol. 2010;21:409-19
9. Abel T1, Zukin RS. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr Opin Pharmacol. 2008;8:57-64
10. Guan JS, Haggarty SJ, Giacometti E. et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55-60
11. MacDonald JL, Roskams AJ. Histone deacetylases 1 and 2 are expressed at distinct stages of neuro-glial development. Dev Dyn. 2008;237:2256-67
12. Yao ZG, Liu Y, Zhang L. et al. Co-location of HDAC2 and insulin signaling components in the adult mouse hippocampus. Cell Mol Neurobiol. 2012;32:1337-42
13. Yoo JY, Larouche M, Goldowitz D. The Expression of HDAC1 and HDAC2 During Cerebellar Cortical Development. Cerebellum. 2013;12:534-46
14. A. M. Baraka, W.H.E.N.S.E.G. Investigating the Role of Zinc in a Rat Model of Epilepsy. CNS Neuroscience & Therapeutics. 2012;18:327-333
15. Dietrich HS, Christian S. Achim Jörres and Joerg C Schefold. Changes in serum creatinine in the first 24 hours after cardiac arrest indicate prognosis: an observational cohort study. Critical Care. 2009;13:1-7
16. Marangos PJ SD, Parma AM, Clark RL, Goodwin FK. Measurement of neuron-specific (NSE) and non-neuronal (NNE) isoenzymes of enolase in rat, monkey and human nervous tissue. J Neurochem. 1979;33:319-29
17. Jiang WYQ, Gong M. et al. Vitamin A deficiency impairs postnatal cognitive function via inhibition of neuronal calcium excitability in hippocampus. J. Neurochem. 2012;121:932-943
18. McHugh M.W.J.a.T.J. Updating hippocampal representations: CA2 joins the circuit. Trends in Neurosciences. 2011;34:526-535
19. Hildmann C RD, Schwienhorst A. Histone deacetylases--an important class of cellular regulators with a variety of functions. Appl Microbiol Biotechnol. 2007;75:487-97
20. Broide RS RJ, Aftahi N, Young W, Bloom FE, Winrow CJ. Distribution of histone deacetylases 1-11 in the rat brain. J Mol Neurosci. 2007;31:47-58
21. Fitzsimons HL, Scott MJ. Genetic modulation of Rpd3 expression impairs long-term courtship memory in Drosophila. PLoS One. 2011;6:e29171
22. Graff J, Rei D, Guan JS. et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483:222-6
23. Hanson JE, Deng L, Hackos DH. et al. Histone deacetylase 2 cell autonomously suppresses excitatory and enhances inhibitory synaptic function in CA1 pyramidal neurons. J Neurosci. 2013;33:5924-9
24. Jawerka M, Colak D, Dimou L. et al. The specific role of histone deacetylase 2 in adult neurogenesis. Neuron Glia Biol. 2010;6:93-107
25. Lubin NMGaFD. The dynamics of HDAC activity on memory formation. Cellscience. 2009;6:44-8
26. Xu K, Dai XL, Huang HC, Jiang ZF. Targeting HDACs: a promising therapy for Alzheimer's disease. Oxid Med Cell Longev. 2011;2011:143269
27. Hawk JD, Florian C, Abel T. Post-training intrahippocampal inhibition of class I histone deacetylases enhances long-term object-location memory. Learn Mem. 2011;18:367-70
28. Montgomery RL, Hsieh J, Barbosa AC, Richardson JA, Olson EN. Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. Proc Natl Acad Sci U S A. 2009;106:7876-81
29. Willis-Martinez D, Richards HW, Timchenko NA, Medrano EE. Role of HDAC1 in senescence, aging, and cancer. Exp Gerontol. 2010;45:279-85
30. Akhtar MW RJ, Nelson ED, Montgomery RL. et al. Histone deacetylases 1 and 2 form a developmental switch that controls excitatory synapse maturation and function. J Neurosci. 2009;29:8288-97
31. Govindarajan N, Rao P, Burkhardt S. et al. Reducing HDAC6 ameliorates cognitive deficits in a mouse model for Alzheimer's disease. EMBO Mol Med. 2013;5:52-63
32. Segre CV, Chiocca S. Regulating the regulators: the post-translational code of class I HDAC1 and HDAC2. J Biomed Biotechnol. 2011;2011:690848
33. Steliou K, Boosalis MS, Perrine SP, Sangerman J, Faller DV. Butyrate histone deacetylase inhibitors. Biores Open Access. 2012;1:192-8
34. Jiang W, Wen EY, Gong M. et al. The pattern of retinoic acid receptor expression and subcellular, anatomic and functional area translocation during the postnatal development of the rat cerebral cortex and white matter. Brain Res. 2011;1382:77-87
35. Mackem S BC, Hager GL. A glucocorticoid/retinoic acid receptor chimera that displays cytoplasmic/nuclear translocation in response to retinoic acid. A real time sensing assay for nuclear receptor ligands. J Biol Chem. 2001;276:45501-4
36. Nakazawa K QM, Chitwood RA, Watanabe M. et al. Requirement for hippocampal CA3 NMDA receptors in associative memory recall. Science. 2002;297:211-8
37. Tsien JZ HP, Tonegawa S. The essential role of hippocampal CA1 NMDA receptor-dependent synaptic plasticity in spatial memory. Cell. 1996;87:1327-38
38. Chandramohan Y, Droste SK, Reul JM. Novelty stress induces phospho-acetylation of histone H3 in rat dentate gyrus granule neurons through coincident signalling via the N-methyl-D-aspartate receptor and the glucocorticoid receptor: relevance for c-fos induction. J Neurochem. 2007;101:815-28
39. Johannes M.H.M. Reul SAH, Andrew Collins and María Gutièrrez Mecinas. Epigenetic mechanisms in the dentate gyrus act as a molecular switch in hippocampus-associated memory formation. Epigenetics. 2009;4:434-9
40. Kee HJ, Kook H. Roles and targets of class I and IIa histone deacetylases in cardiac hypertrophy. J Biomed Biotechnol. 2011;2011:928326
Author contact
![]() Corresponding author: Children Nutrition Research Center, Children's Hospital of Chongqing Medical University, Chongqing 400014, China; Tel: +86 23 63630913; Fax: +86 23 63622754; Email: tylisina.com (Tingyu Li) or jchen010com (Jie Chen).
Corresponding author: Children Nutrition Research Center, Children's Hospital of Chongqing Medical University, Chongqing 400014, China; Tel: +86 23 63630913; Fax: +86 23 63622754; Email: tylisina.com (Tingyu Li) or jchen010com (Jie Chen).
Received 2013-12-22
Accepted 2014-5-14
Published 2014-5-30
