ISSN: 1449-1907International Journal of Medical Sciences
Int J Med Sci 2013; 10(12):1602-1607. doi:10.7150/ijms.6526 This issue Cite
Research Paper
Immunostaining of p16INK4a/Ki-67 and L1 Capsid Protein on Liquid-based Cytology Specimens Obtained from ASC-H and LSIL-H Cases
1. Department of Obstetrics and Gynecology, The Catholic University of Korea, Seoul, Korea;
2. Department of Hospital Pathology, The Catholic University of Korea, Seoul, Korea.
Abstract
Background: Atypical squamous cell cannot exclude high-grade squamous intraepithelial lesion (ASC-H) and low-grade intraepithelial lesion cannot exclude high-grade squamous intraepithelial lesion (LSIL-H) are ambiguous diagnostic entities for the prediction of high-grade cervical lesion. Objective and reproducible tests for predicting high-grade cervical lesions are needed to reduce unnecessary colposcopic referrals or follow-ups.
Objective: We aimed to identify an adequate set of adjunctive markers to predict cervical intraepithelial neoplasia grade 2+ (CIN2+) in residual liquid-based cytology specimens (LBCS).
Methods: We conducted p16 INK4a/Ki-67 and L1 capsid protein immunostaining and human papillomavirus (HPV) DNA typing on 56 LBCS diagnosed with ASC-H or LSIL-H, all of which were subjected to histologic confirmation or follow-up cytologic examination.
Results: Positivity for p16 INK4a/Ki-67 was associated with a histology of CIN2+ (P=0.047) and CIN3+ (P=0.002). Negativity for L1 capsid protein was associated with CIN2+ confirmed at follow-up (P=0.02).Positivity for high-risk HPV (HR-HPV) was associated with CIN2+ confirmed at follow-up (P=0.036) and a histology of CIN2+ (P=0.037). The sensitivity, specificity, positive predictive value, and negative predictive value for predicting follow-up CIN2+ were 76.2%, 51.4%, 48.5%, and 78.3%, respectively, for p16 INK4a/Ki-67 immunostaining; 95.2%, 34.3%, 46.5%, and 92.3%, respectively, for L1 capsid protein; and 66.7%, 67.7%, 54.5%, and 77.8%, respectively, for HR-HPV. The classification and regression tree analysis showed that the combined results of p16 INK4a/Ki-67 andL1 capsid protein immunostaining and the HR-HPV test, conducted sequentially, correctly classified 81.8% of samples (27/33)in the prediction of a histology of CIN2 + in ASC-H or LSIL-H. For determination of the histology of cervical intraepithelial neoplasia grade 3+ (CIN3+)in ASC-H or LSIL-H, we found that the combined results of p16 INK4a/Ki-67 and L1 capsid protein immunostaining correctly classified 78.8% (26/33) of samples.
Conclusions: p16INK4a/Ki-67 and L1 capsid protein immunostaining and HR-HPV testing of residual LBCS diagnosed with ASC-H or LSIL-H are useful objective biomarkers for predicting CIN2+. Immunostaining for p16INK4a/Ki-67 and L1 capsid protein are sufficient to predict CIN3+.
Keywords: p16 INK4a/Ki-67 dual staining, L1 capsid protein, Human papillomavirus, Cervicovaginal cytology, Immunocytochemistry, HPV DNA typing.
INTRODUCTION
Carcinoma of the uterine cervix is the second most common malignancy in women worldwide [1]. Since the introduction of screening using cervicovaginal cytology, the incidence and mortality of invasive cervical cancer have declined dramatically [2].Human papillomavirus (HPV) infection is necessary for cervical cancer development, but most HPV infections are transient and asymptomatic. Ninety percent of HPV infections disappear within 2 years [3,4]. Currently, neither cervicovaginal cytology nor HPV testing can provide accurate information about which precursor lesions would progress toward cancer [5,6]. In particular, the grey zones of cervical cytological diagnoses such as atypical squamous cell of undetermined significance (ASCUS), atypical squamous cell cannot exclude high-grade squamous intraepitheal lesion (ASC-H), and low-grade squamous intraepithelial lesion cannot exclude high-grade squamous intraepithelial lesion (LSIL-H) cause diagnostic and therapeutic difficulties. LSIL-H shows features of both low-grade squamous intraepithelial lesion (LSIL) and ASC-H, containing both definite low-grade dysplastic squamous cells and a few atypical cells, suspicious but not diagnostic for high-grade squamous intraepitheal lesion (HSIL) [7]. The clinical significance of LSIL-H lies between LSIL and ASC-H [7].
This study analyzed the expression of p16 INK4a/Ki-67 and L1 capsid protein and conducted HPV DNA typing in liquid-based cytology specimens (LBCS) of ASC-H and LSIL-H patients to develop a more effective set of surrogate markers for the prediction of high-risk precursor or invasive cervical lesions.
MATERIAL AND METHODS
Specimens
A total of 56 SurePath®LBCS from 26 cases classified as ASC-H and 30 cases classified as LSIL-H were collected from Seoul St. Mary Hospital between January 2011 and January 2012. Thirty-eight of 56 cases (67.9%) were confirmed by histological examination (Table 1) and the remaining 18 cases were followed up for 4-12 months (mean, 8 months). Additionally, 8 specimens from cases classified as LSIL and 8 specimens from cases of tissue-confirmed HSIL were collected. All histological slides were secondarily reviewed by one pathologist (AWL). The histopathological results showed 12 cases of chronic cervicitis, 8 cases of cervical intraepithelial neoplasia grade 1 (CIN1), 5 cases of cervical intraepithelial neoplasia grade 2 (CIN2), and 23 cases of cervical intraepithelial neoplasia grade 3 (CIN3) (Table 1). Two additional unstained SurePath® slides were prepared from each specimen for immunocytochemical staining, and 1.0 mL of residual material was collected for the HPV DNA test. Histologically confirmed ≥CIN2 was considered “Histology CIN2+”; histologically confirmed ≥CIN3 was considered “Histology CIN3+”; and histologically confirmed ≥CIN2+ or cytologically diagnosed ≥HSIL at follow-up was considered “Follow-up CIN2+.”
This study was approved by the ethical committees of the individual institutions (IRB approval number KC12SISI0573).
Cervical Cytology versus Biopsy Results.
| Biopsy results | |||||
|---|---|---|---|---|---|
| Cytology results | NILM | CIN I | CIN II | CIN III | Total |
| LSIL | 1 | 1 | 0 | 0 | 2 |
| LSIL-H | 8 | 2 | 2 | 3 | 15 |
| ASC-H | 3 | 5 | 2 | 13 | 23 |
| HSIL | 0 | 0 | 1 | 7 | 8 |
| Total | 12 | 8 | 5 | 23 | 48 |
LILM = negative for intraepithelial lesion or malignancy.
CIN = cervical intraepithelial neoplasm. LSIL = low grade squamous intraepithelial lesion.
LSIL-H = low grade intraepithelial lesion cannot exclude HSIL. ASC-H = atypical squamous cell cannot exclude HISL.
HSIL = high grade squamous intraepithelial lesion.
Immunocytochemical staining for p16 INK4a/Ki-67
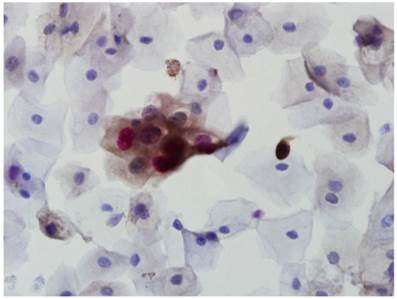
In order to detect overexpression of p16 INK4a/Ki-67, immunocytochemistry was performed using the CINtec® PLUS kit (MTM Laboratories, Heidelberg, Germany) according to the manufacturer's protocol [8]. A p16 INK4a/Ki-67-positive result was defined as the detection of simultaneous co-localization of p16 INK4a and Ki-67 staining in the same cervical epithelial cell (Fig. 1).
CINtec PLUS double-positive cells showing both brown cytoplasmic staining for p16 and red nuclear staining for Ki-67 (×400).
Immunocytochemical staining for HPV L1 capsid protein
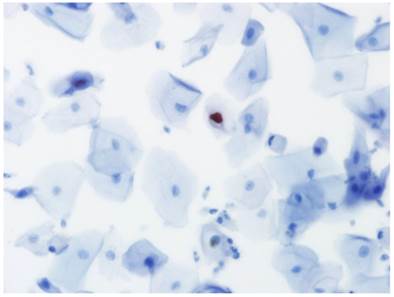
Immunocytochemistry for the presence of L1 capsid protein was performed using the Cytoactive® HPV L1 screening set (Cytoimmun Diagnostics GmbH, Pirmasens, Germany) according to manufacturer's protocols [9]. The L1 antibody recognizes the major L1 capsid protein of 28 HPV types including types 6, 11, 16, 18, 31, and 45. Specimens incubated with phosphate-buffered saline (PBS) instead of primary antibody were used as negative controls; specimens confirmed to be condyloma by biopsy were used as positive controls. In the evaluation of L1 immunocytochemical staining, only nuclear staining was considered positive (Fig. 2).
Positive staining for HPV L1 capsid protein in the nucleus of an epithelial cell in liquid-based cytology (×400).
The PANArray™ HPV chip test
The PANArray™ HPV chip test (Panagene, Daejeon, Korea) detects 19 high-risk (16, 18, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 70 26, 69, and 73) and 13 low-risk (6, 11, 34, 40, 42, 43, 44, 54 32, 55, 62, 81, 83) HPV types. Analyses were performed according to the manufacturer's instructions [10].
Statistical analysis
All statistical analyses were performed using SPSS version 13.0 (SPSS Inc., Chicago, IL) for Windows. The chi-square test was used to determine an association between the results of analyses for p16 INK4a/Ki-67, L1 capsid protein, high-risk HPV (HR-HPV) and 16,18-HPV and those of follow-up CIN2+, histology CIN2+ and histology CIN3+, respectively. Diagnostic sensitivities, specificities, positive predictive values (PPV), and negative predictive values (NPV) were calculated. A CIN2+ or CIN3+ prediction tree was constructed using the classification and regression tree (CRT) method. A P-value <0.05 was considered significant.
RESULTS
The patients ranged in age from 25 to 83 years, with a mean age of 46 years and a median age of 45 years.
The p16 INK4a/Ki-67 immunocytochemistry
Among the 56 cases with cytological diagnoses of ASC-H or LSIL-H, 33 cases (58.9%) were positive for p16 INK4a/Ki-67. Sixteen of the 21 follow-up CIN2+ cases (76.2%) and 15 of the 20 histology CIN2+ cases (75%) were positive for p16 INK4a/Ki-67 (Tables 2 and 3). Two of the 8 LSIL cases (25%) and 6 of the 8 HSIL cases (75%) were positive for p16 INK4a/Ki-67. Positivity for p16 INK4a/Ki-67 was associated with histology CIN2+ (P=0.047) and histology CIN3+ (P=0.002). Positivity for p16 INK4a/Ki-67 was not associated with follow-up CIN2+ confirmation (P=0.053) (Tables 2 and 3).
The L1 capsid protein immunocytochemistry
Among the 56 cases with cytological diagnoses of ASC-H or LSIL-H, 13 (23.2%) were positive for L1 capsid protein. One of the 21 follow-up CIN2+ cases (4.8%) and 1 of the 20 histology CIN2+ cases (5%) were positive for L1 capsid protein (Tables 2 and 3). Three of the 8 LSIL cases (37.5%) and 1 of the 8 HSIL cases (12.5%) were positive for L1 capsid protein. Negativity for L1 capsid protein was associated with follow-up CIN2+ (P=0.02) but not with histology CIN2+ (P=0.083) or histology CIN3+ (P=0.370) (Tables 2 and 3).
The PANArray™ HPV chip test
Among the 49 cases with cytological diagnoses of ASC-H or LSIL-H tested, 22 (44.9%) were positive for HR-HPV. Of the 22 HR-HPV cases, 11 were HPV 16 and/or HPV 18 and the remaining 11 were non-16,18 HR-HPV. The HPV chip test was not performed for 7 of 56 cases (12.5%) due to shortage of residual samples. Twelve of the 18 follow-up CIN2+ cases (66.7%) and 12 of the 18 histology CIN2+ cases (66.7%) were positive for HR-HPV (Tables 2 and 3).Two of the 8 LSIL cases (25%) and 4 of the 7 HSIL cases (57.1%) were positive for HR-HPV. Positivity for HR-HPV was associated with follow-up CIN2+ (P=0.036) and histology CIN2+ (P=0.037).Positivity for HR-HPV was not associated with CIN3+ (P=0.491) (Tables 2 and 3).Of the 18 follow-up CIN2+ cases, 7 (15.1%) were positive for 16,18-HPV;of the 14 histology CIN3+ cases, 5 (35.7%) were positive for 16,18-HPV (Tables 2 and 3).Two of the 8 LSIL cases (25%) and none of the 7 HSIL cases (0%) were positive for 16,18-HPV. Positivity for 16,18-HPV showed no association with follow-up CIN2+ (P=0.072), histology CIN2+ (P=0.134), or histology CIN3+ (P=0.442) (Tables 2 and 3).
Correlation of the results of p16 INK4a/Ki-67 and L1 capsid protein immunostaining and HPV DNA typing with follow-up CIN2+.
| Follow up results | P value | |||
|---|---|---|---|---|
| <CIN2 | ≥CIN2 | |||
| p16 INK4a /Ki-67 (n=56) | positive | 17 | 16 | 0.053 |
| negative | 18 | 5 | ||
| L1 capsid protein (n=56) | positive | 12 | 1 | 0.02 |
| negative | 23 | 20 | ||
| HR-HPV (n=49) | positive | 10 | 12 | 0.036 |
| negative | 21 | 6 | ||
| 16,18-HPV (n=49) | positive | 4 | 7 | 0.072 |
| negative | 27 | 11 | ||
CIN = cervical intraepithelial neoplasm.
HR-HPV = high risk type HPV.
16,18-HPV = HPV type 16 and/or HPV type 18.
Correlation of the results of p16 INK4a/Ki-67 and L1 capsid protein immunostaining and HPV DNA typing with histology CIN2+ and histology CIN3+.
| Histologic results | |||||||
|---|---|---|---|---|---|---|---|
| <CIN2 | ≥CIN2 | P value | <CIN3 | ≥CIN3 | P value | ||
| p16 INK4a /Ki-67 (n=38) | pos | 7 | 15 | 0.047 | 8 | 14 | 0.002 |
| neg | 11 | 5 | 14 | 2 | |||
| L1 capsid protein (n=38) | pos | 5 | 1 | 0.083 | 5 | 1 | 0.370 |
| neg | 13 | 19 | 17 | 15 | |||
| HR-HPV (n=33) | pos | 4 | 12 | 0.037 | 8 | 8 | 0.491 |
| neg | 11 | 6 | 11 | 6 | |||
| 16,18-HPV (n=33) | pos | 2 | 7 | 0.134 | 4 | 5 | 0.442 |
| neg | 13 | 11 | 15 | 9 | |||
CIN = cervical intraepithelial neoplasm. HR-HPV = high risk type HPV. 16,18-HPV = HPV type 16 and/or HPV type 18.
Diagnostic efficiency to predict CIN2+ by follow-up CIN2+ in ASC-H and LSIL-H
The sensitivity, specificity, PPV, and NPV for predicting follow-up CIN2+ were 76.2%, 51.4%, 48.5%, and 78.3%, respectively, for p16 INK4a/Ki-67; 95.2%, 34.3%, 46.5%, and 92.3%, respectively, for L1 capsid protein;66.7%, 67.7%, 54.5%, and 77.8%, respectively, for HR-HPV; and38.9%, 87.1%, 63.6%, and 71.1%, respectively, for 16,18-HPV (Table 4). The sensitivity, specificity, PPV, and NPV of the combined results of p16 INK4a/Ki-67 and L1 capsid protein (p16 INK4a/Ki-67, L1 capsid protein (+/-) vs. other result) were 61.9%, 68.6%, 54.2%, and 75.0%, respectively. When the HR-HPV test was positive, or p16 INK4a/Ki-67 and L1 capsid protein immunostaining were positive and negative, respectively, the sensitivity, specificity, PPV, and NPV of this set were 100%, 53.1%, 55.9%, and 100%, respectively.
Diagnostic effectiveness of p16 INK4a/Ki-67 immunocytostaining, L1 capsid protein immunocytostaining, and PANArray™ HPV chip test to predict follow-up CIN2+.
| Sensitivity(%) | Specificity(%) | PPV(%) | NPV(%) | |
|---|---|---|---|---|
| p16 INK4a /Ki-67 | 76.2 | 51.4 | 48.5 | 78.3 |
| L1 capsid protein | 95.2 | 34.3 | 46.5 | 92.3 |
| HR- HPV | 66.7 | 67.7 | 54.5 | 77.8 |
| 16,18-HPV | 38.9 | 87.1 | 63.6 | 71.1 |
| p16 /Ki-67 + L1 | 61.9 | 68.6 | 54.2 | 75.0 |
| p16 /Ki-67 + L1 + HR-HPV | 100 | 53.1 | 55.9 | 100 |
PPV = positive predictive value. NPV = negative predictive value.
HR-HPV = positive for high risk HPV.
16,18-HPV =positive for HPV type 16 or HPV type 18.
p16/Ki-67 + L1 = positive for p16 INK4a/Ki-67 and negative for L1 capsid protein.
p16/Ki-67 + L1 + HR-HPV = (positive for p16 INK4a/Ki-67 and negative for L1 capsid protein) or (positive for high risk HPV).
Classification and regression tree analysis for predicting a histology CIN2+ and histology CIN3+ in ASC-H and LSIL-H cases
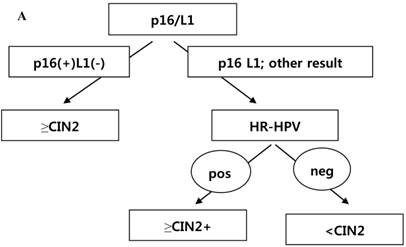
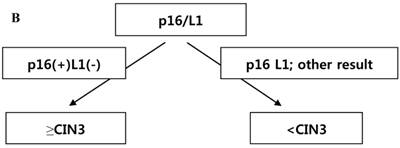
CRT analysis was performed to predict histology CIN2+ and histology CIN3+ in cases with the diagnosis of ASC-H or LSIL-H. The independent variables included were p16 INK4a/Ki-67, L1 capsid protein, HR-HPV,16,18-HPV, and the combined results of p16 INK4a/Ki-67 and L1 capsid protein. To predict histology CIN2+, the primary variable for hierarchical tree was the combined results of p16 INK4a/Ki-67 and L1 capsid protein, and the secondary variable was HR-HPV (Fig. 3A). However, to predict histology CIN3+, the applied variable was only the combined results of p16 INK4a/Ki-67 and L1 capsid protein (Fig. 3B). Using the CRT analysis, 81.8% (27/33) and 78.8% (26/33) of cases were correctly classified as histology CIN2+ and histology CIN3+, respectively.
DISCUSSION
Since HPV is a definitive etiological agent of cervical cancer, HPV testing can be a potentially important tool in cervical cancer screening programs. However, the HPV test cannot differentiate clinically relevant infections from transient infections that do not lead to cervical neoplasia. As a result, complementary biomarkers are needed to allow such discrimination.
(A) Classification and regression tree analysis to predict histology CIN2+. p16 = p16INK4a/Ki-67. L1 = L1 capsid protein. HR-HPV =high risk type HPV. (B) Classification and regression tree analysis to predict histology CIN3+. p16 = p16INK4a/Ki-67. L1 = L1 capsid protein.
The p16INK4a protein inhibits the cyclin-dependent kinases, which regulate progression through the cell cycle by phosphorylating the retinoblastoma protein (pRb). It has been demonstrated that p16INK4a accumulation is involved in a negative feedback loop with pRb. Hence, the decreased Rb function enhances p16INK4a expression, which can precisely indicate the degree of the HPV-related cervical epithelial lesion [11].
Sahebaliet al. [12] reported that Ki-67 immunocytochemistry can be applied to LBC and that it yields significant positive staining results in HSIL- and HPV-16-positive samples. The use of a dual immunocytochemistry technique that is specific for the detection of the cyclin-dependent kinase inhibitor p16 and the proliferation marker Ki-67 may be useful for such purposes. In fact, p16 overexpression suggests the presence of the viral oncoprotein E7, and the concurrent presence of Ki-67 suggests altered cell cycle control. Therefore, the simultaneous expression of both markers is suggestive of a transforming infection.
Positive immunostaining of p16 INK4a/Ki-67 significantly increased with the severity of the cytological and histological abnormalities. Positive p16 INK4a/Ki-67 immunostaining had a strong association with a CIN2+ diagnosis (COR = 10.86; 95% CI, 4.16-29.12)[13]. These results were reaffirmed in the current study: positive p16 INK4a/Ki-67 immunostaining results were associated with histology CIN2+ and histology CIN3+ (P=0.047 and P=0.002, respectively). The clinical performance of p16 INK4a/Ki-67dual staining has been evaluated in 2 studies, where it was used as a triage test [14,15]. One study demonstrated 91.9% sensitivity of dual-stain testing for the detection of biopsy-confirmed CIN2+ during preliminary follow-up within the group of Pap-negative/HPV-positive women, and 82.1% specificity for CIN2+ on biopsy [14].The other study noted 92.2% (in ASCUS cases) and 94.2% (in LSIL cases) sensitivity, and 80.6% (in ASCUS cases) and 94.2% (in LSIL cases) specificity, of p16 INK4a/Ki-67dual-stain cytology for biopsy-confirmed CIN2+. Dual-stain cytology showed comparable sensitivity, but significantly higher specificity as compared to HPV testing [15]. In the current study, the p16 INK4a/Ki-67 immunostaining showed similar sensitivity (76.2%) but lower specificity (51.4%) than the previous study.
Most previous studies, including the 2 clinical studies of p16 INK4a cited above, were performed on specimens prepared with a ThinPrep®kit (Hologic, Inc., Marlborough, MA), which uses a different fixative media and slide-preparation method as opposed to a SurePath®kit (BD Diagnostics-Ripath, Burlington, NC). A retrospective study performed using SurePath® preparations to evaluate the performance of p16 INK4a/Ki-67 immunostaining in cases diagnosed as ASCUS showed similar specificity (53%) and sensitivity (64%) as that of the current study in the prediction of high-grade disease at surgical biopsy [16]. The authors of that study suggested that the differences in LBCS preparation and the lack of a secondary review and reinterpretation of discrepant cases may have led to lower sensitivity [16]. In this study, we used SurePath®-prepared specimens and did not re-evaluate the cytological or immunocytochemical results in discrepant cases to reflect a real practice setting.
L1 capsid protein, the major structural protein of HPV, is expressed in the early productive phase of cervical cancer but is gradually lost in the later proliferative phase when HPV DNA is integrated into the host DNA [17]. In the current study, L1 capsid protein negativity in LBCS correlated with follow-up CIN2+ (P=0.02). This result is consistent with a study in which L1 capsid protein expression in cervical cytology negatively correlated with cervical lesion severity [9].
When we combined the results of p16 INK4a/Ki-67 and L1 capsid protein (p16 INK4a/Ki-67, L1 capsid protein (+/-) vs. other results), the diagnostic accuracy of the combined results was similar to that of HR-HPV testing (Table 4). In the present study, when we obtained either a positive HR-HPV result or a p16 INK4a/Ki-67 and L1 capsid protein (+/-) result, we predicted a follow-up CIN2+ with 100% sensitivity and 100% NPV, but with low specificity (53.1%) and low PPV (55.9%). With this high sensitivity and NPV, clinicians may be able to reduce unnecessary colposcopic referrals or follow-up in ASC-H and LSIL-H cases, but a larger study would be necessary to confirm this finding.
To predict a histology of CIN2+, a hierarchical tree using CRT analysis was made. The primary variable for the hierarchical tree was the combined results of p16 INK4a/Ki-67 and L1 capsid protein immunostaining, and the secondary variable applied was the result of the HR-HPV test. To predict CIN3+, the combined results of p16 INK4a/Ki-67 and L1 capsid protein immunostaining were the only variables applied (Fig. 3A and 3B).When we followed this tree, we could correctly classify a histology of CIN2+ and histology of CIN3+ in 81.8% and78.8% of cases, respectively. This tree method has not previously been used to predict ≥CIN2 and ≥CIN3.
In conclusion, the combination of p16INK4a/Ki-67 andL1 capsid protein immunostaining and HR-HPV DNA testing of residual LBCS provides useful objective biomarkers to predict CIN2+ in cases diagnosed as ASC-H or LSIL-H; p16INK4a/Ki-67 and L1 capsid protein immunostaining are sufficient to predict CIN3+.
Abbreviations
ASC-H: Atypical squamous cell cannot exclude HISL; ASCUS: Atypical squamous cell of undetermined significance; CIN: Cervical intraepithelial lesion; CRT: classification and regression tree; HPV: human papillomavirus; HR-HPV: High risk type HPV; HSIL: High grade squamous intraepitheal lesion; LBCS: Liquid-based cytology specimens; LSIL: Low grade squamous intraepithelial lesion; LSIL-H: Low grade intraepithelial lesion cannot exclude HSIL; NPV: Negative predictive value; PPV: Positive predictive value; 16,18-HPV: HPV type 16 and/or HPV type 18.
Acknowledgements
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2011-0027250).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Parkin DM, Pisani P, Ferlay J. Estimates of the worldwide incidence of eighteen major cancers in 1985. Int J Cancer. 1993;54:594-606
2. Denny L. The prevention of cervical cancer in developing countries. BJOG. 2005;112:1204-12
3. Walboomers JM, Jacobs MV, Manos MM. et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12-9
4. Steben M, Duarte-Franco E. Human papillomavirus infection: epidemiology and pathophysiology. Gynecol Oncol. 2007;107:S2-5
5. Ostör AG. Natural history of cervical intraepithelial neoplasia: a critical review. Int J Gynecol Pathol. 1993;12:186-92
6. Klaes R, Benner A, Friedrich T. et al. p16INK4a immunohistochemistry improves interobserver agreement in the diagnosis of cervical intraepithelial neoplasia. Am J Surg Pathol. 2002;26:1389-99
7. Elsheikh TM, Kirkpatrick JL, Wu HH. The significance of “low-grade squamous intraepithelial lesion, cannot exclude high-grade squamous intraepithelial lesion” as a distinct squamous abnormality category in papanicolaou tests. Cancer. 2006;25:277-81
8. Yoshida T, Sano T, Kanuma T. et al. Usefulness of CINtec® PLUS p16/Ki-67 double-staining in cytological screening of cervical cancer. Acta Cytol. 2011;55:413-20
9. Huang MZ, Li HB, Nie XM. et al. An analysis on the combination expression of HPV L1 capsid protein and p16INK4a in cervical lesions. Diagn Cytopathol. 2010;38:573-8
10. Choi JJ, Kim C, Park H. Peptide nucleic acid-based array for detecting and genotyping human papillomaviruses. J Clin Microbiol. 2009;47:1785-90
11. Kim YT, Zhao M. Aberrant cell cycle regulation in cervical carcinoma. Yonsei Med J. 2005;46:597-613
12. Sahebali S, Depuydt CE, Segers K. et al. Ki-67 immunocytochemistry in liquid based cervical cytology: useful as an adjunctive tool?. J Clin Pathol. 2003;56:681-6
13. Donà MG, Vocaturo A, Giuliani M. et al. p16/Ki-67 dual staining in cervico-vaginal cytology: correlation with histology, human papillomavirus detection and genotyping in women undergoing colposcopy. Gynecol Oncol. 2012;126:198-202
14. Petry KU, Schmidt D, Scherbring S. et al. Triaging Pap cytology negative, HPV positive cervical cancer screening results with p16/Ki-67 dual-stained cytology. Gynecol Oncol. 2011;121:505-9
15. Schmidt D, Bergeron C, Denton KJ, Ridder R. p16/Ki-67 dual-stain cytology in the triage of ASCUS and LSIL papanicolaou cytology: results from the European equivocal or mildly abnormal Papanicolaou cytology study. Cancer Cytopathol. 2011;119:158-66
16. Edgerton N, Cohen C, Siddiqui MT. Evaluation of CINtec PLUS® testing as an adjunctive test in ASC-US diagnosed SurePath® preparations. Diagn Cytopathol. 2013;41:35-40
17. Hilfrich R, Hariri J. Prognostic relevance of human papillomavirus L1 capsid protein detection within mild and moderate dysplastic lesions of the cervix uteri in combination with p16 biomarker. Anal Quant Cytol Histol. 2008;30:78-82
Author contact
![]() Corresponding author: Ahwon Lee, M.D., Ph.D. Department of Hospital Pathology, The Catholic University of Korea, Seoul St. Mary's Hospital, 505 Banpo-Dong, Seocho-Gu, Seoul137-040, Korea. Tel: +82-2-2258-1621, Fax: +82-2-2258-1627 E-mail: kleeac.kr.
Corresponding author: Ahwon Lee, M.D., Ph.D. Department of Hospital Pathology, The Catholic University of Korea, Seoul St. Mary's Hospital, 505 Banpo-Dong, Seocho-Gu, Seoul137-040, Korea. Tel: +82-2-2258-1621, Fax: +82-2-2258-1627 E-mail: kleeac.kr.
Received 2013-4-21
Accepted 2013-8-21
Published 2013-9-12
